Abstract
Purpose
The objective of this experimental study was to compare the global gene expression profile of CC of mature oocytes in 18 patients with severe endometriosis and CC in 18 control patients affected by a severe male factor.
Methods
For each group, the CC were pooled, RNA was extracted and a microarray performed. For validating the microarray, a quantitative real-time PCR was performed in the CC of an independent set of patients with endometriosis (n = 5) and controls (n = 7).
Results
595 differentially expressed genes (320 down-regulated, 275 up-regulated, p < 0.05, fold change ≥1.5) were identified. The most significant changes were observed in genes involved in the chemokine signaling and cell-cell or cell-extracellular matrix adhesion pathways. Several genes of these pathways were down-regulated in endometriosis. Individual RT-PCR assays confirmed the microarray for ten genes.
Conclusions
Several genes involved in the chemokine mediated-signaling pathway and in the functional cross-talk between CC and the oocyte are down-regulated in endometriosis CC. The impairment of these processes could explain the reduction of oocyte competence in endometriosis. This preliminary knowledge could be the starting point for a more detailed elucidation of the relationship between endometriosis and oocyte competence.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-014-0305-1) contains supplementary material, which is available to authorized users.
Keywords: Endometriosis, Cumulus cells, Microarray, IVF
Introduction
The effect of endometriosis on assisted reproductive treatments is controversial. A milestone meta-analysis of observational studies has demonstrated that patients with endometriosis-associated infertility have shown a decrease in the number of retrieved oocytes, in fertilization, implantation, and pregnancy rates, in comparison with patients affected by tubal factor infertility [1]. Furthermore, pregnancy rates in patients with severe endometriosis were significantly lower than those of patients with mild disease [1]. Based on the results of this meta-analysis, in 2005 ESHRE guidelines for the diagnosis and treatment of endometriosis [2] have indicated a high level of evidence with grade A strength for the reduction of pregnancy rates in patients with endometriosis, compared to those with tubal infertility. In 2012, the Practice Committee of the American Society for Reproductive Medicine [3] concurred with this conclusion.
In a large database-cohort study, decreasing fertilization and pregnancy rates were observed in patients with endometriosis, compared to patients with other causes of infertility [4]. Nevertheless, the findings of these studies contrast with data coming from the registry of the Society for Assisted Reproductive Technology (SART). Here, patients with endometriosis shared similar implantation, pregnancy and delivery rates, compared with those patients affected by other causes of infertility [5]. The discrepancy between the first two studies [1, 4] and the last [5] may be due to the fact that data from national registries are not adjusted for confounding variables. Taking into account these last data, the new ESHRE guidelines [6] did not indicate the same level of evidence as the previous guidelines [2] for the reduction of pregnancy rates in patients with endometriosis. Furthermore, other clinical studies have demonstrated a major impairment in oocyte quality in patients with endometriosis. A study carried out on oocyte donors revealed that recipients with endometriosis did not show a reduced pregnancy rate. On the contrary, the oocytes retrieved by donors with endometriosis led to a statistically significant decrease in pregnancy rates in recipients, thereby demonstrating a direct effect of endometriosis on oocyte and embryo quality rather than on endometrial receptivity [7]. The results of this study have been confirmed by other observations, suggesting that an altered follicular microenvironment in patients with endometriosis could be responsible for a defective oogenesis and subsequently low fertilization rates or poor quality embryos with a reduced ability to implant [8].
In order to evaluate oocyte competence, a non-invasive method has been proposed, involving the gene expression analysis of cumulus cells (CC). These cells surround the oocyte and establish a functional bidirectional cross-talk with the oocyte through gap junctions and paracrine factors. Several studies have been published, demonstrating that the gene expression of CC may differ in relation to the potential development of the embryo [9–15]. Furthermore, a relationship between the kind of stimulation during an IVF cycle and differences in the gene expression profile of CC has been demonstrated [9, 15–17].
As far as is known, no genome-wide expression study on CC of mature oocytes in patients affected by severe endometriosis has been published. The available evidence supports the thesis that modifications in granulosa cells may impair follicular growth and oocyte quality in patients with endometriosis [18]. Increased apoptotic activity, related to the severity of the disease, has been described in the granulosa cells of endometriosis patients, resulting in poor oocyte quality and a reduction in fertilization and pregnancy rates [19]. Commencing with these initial observations, the objective of this study was a comparison of the global gene expression profile of CC, associated with mature oocytes isolated from patients affected by severe endometriosis, and from control patients referred for in vitro fertilization for a severe male factor.
Material and methods
Patient population and ovarian stimulation
In this experimental study, CC obtained from 18 patients attending an intracytoplasmic sperm injection (ICSI) cycle for severe endometriosis (Group A) were compared to control CC obtained from 18 patients attending an ICSI cycle for severe male factor (Group B) at the ANDROS Day Surgery Clinic Reproductive Medicine Unit in Palermo, Italy. The patients with endometriosis underwent ICSI in order to have two groups treated in the same way, avoiding any bias. The diagnosis of severe endometriosis (stage III-IV of the ASRM revised classification) [20] was performed by laparoscopy or laparotomy and histologically confirmed. No patient had an ovarian endometrioma at the moment of enrollment to the study.
Severe male factor infertility was defined as a sperm count ≤5.000.000/ml or azoospermia. The inclusion criteria were as follows:
the patients were at least 18 years of age but not >38 years
they had a BMI of between 18 and 30 kg/m2 and
normal regular menstrual cycles, ranging from 24 to 35 days in length, basal serum FSH ≤12 mIU/mL, basal AMH level between 0.6 and 6 ng/ml, normal thyroid-stimulating hormone (TSH) and prolactin levels, and normal uterine cavity as assessed by hysteroscopy.
Those patients affected by polycystic ovary syndrome or other anovulatory causes of infertility were excluded. Controlled ovarian hyperstimulation was performed after pituitary down-regulation with a gonadotrophin-releasing hormone agonist (GnRH-a, leuprolide acetate; 3.75 IM) (Enantone; Takeda Chemical Industries, Osaka, Japan), administered on day 21 of the previous cycle. Multifollicular development was achieved by daily injections of recombinant FSH (Gonal F; Serono, Rome, Italy), commencing after at least 12 days of pituitary down-regulation.
Treatment with a fixed dose of recombinant FSH was initiated for five days and then adjusted, according to the response of the ovaries to stimulation. Oocyte maturation was triggered with an injection of 10,000 IU of human chorionic gonadotropin (hCG, Gonasi HP; IBSA, Switzerland) after at least 10 days of FSH therapy and when three follicles reached 16 mm. The oocyte pick-ups were carried out 36 h after the triggering of oocyte maturation. ICSI and embryo transfer procedures were performed according to the protocols described in Volpes et al., 2004 [21]. The study was approved by the local Ethical Committee (the Ethical Committee of the ANDROS Day Surgery Clinic, Palermo, approval date 22/01/2010, Cod. 01/MR/10).
Isolation and collection of cumulus cells
After oocyte pick up, every oocyte surrounded by its own CC was cultured individually in 30 μl microdrops of culture media (ISM1, Origio) under mineral oil (Liquid Paraffin, Origio) for at least two hours [22]. Decoronization was carried out by placing each cumulus oocyte complex (COC) in separated drops of recombinant human hyaluronidase (ICSI cumulase, Origio) under oil. Sterile plastic strippers were used (Stripper Tips, Origio) to separate the oocyte from the cumulus cells.
The oocyte was then washed in 30 μl microdrops of a HEPES buffered medium (Flushing Medium, Origio) under oil, and nuclear maturation was evaluated by using an inverted microscope at 400 magnification. The collected oocytes were classified as follows: mature oocytes (MII), immature oocytes (MI and germinal vesicles GV). Drops containing CC from immature oocytes were discarded.
All the cumulus cells associated with mature oocytes were aspirated from drops of recombinant human hyaluronidase for each patient, pooled and placed into a conical plastic tube containing 10 ml of sterile saline solution. The tube was centrifuged, the supernatant was discarded and the pellet was re-suspended in 1 ml of fresh sterile saline solution. A second centrifugation was performed by using a 1 ml conical Eppendorf tube. The supernatant was discarded, the pellet was re-suspended in 0.5 ml of RNAlater solution (Ambion, TX, USA) and the Eppendorf tube was cryo-preserved at −80 °C.
RNA isolation and microarray analysis
Prior to RNA extraction, oophorus cumuli from 18 endometriosis patients and from 18 control patients were pooled. A pooled sample containing 77 oophorus cumuli was obtained for the endometriosis group (group A), and 110 oophorus cumuli were collected from the control group (group B). Total RNA was extracted from CC, according to the manufacturer’s protocol (Affymetrix, Santa Clara, CA). Quantity and purity were determined for all RNA samples using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies Celbio, Italy) and RNA integrity was assessed using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). A microarray analysis was performed, according to the manufacturer’s instructions (Affymetrix, Santa Clara, CA) by using the Two-Cycle Target Labeling and Control Reagents kit (Affymetrix, Santa Clara, USA). The two-cycle labeling protocol consisted of two successive rounds of T7-based in vitro transcription, incorporating biotin rNTPs in the second round reaction. Fifty nanograms of total RNA were amplified using the two-cycle cDNA synthesis kit (Affymetrix, Santa Clara, USA), in combination with the MEGA script T7 in vitro transcription system (Ambion, TX, USA). Two biological replicates were performed for each experimental condition. AffymetrixGeneChip Operating Software (Affymetrix GCOS v1.4) was used to analyze the image data. The quality control of the arrays was performed using AffyQCReport software [23] and comparable quality between probe arrays was demanded for all arrays in each experiment.
Heat Map analyses (HCA) were performed using the MultiExperiment Viewer (MeV v4.8) program from the TM4 Microarray Software Suite. An Agene Set Analysis Toolkit was used to investigate the biological significance of a set of genes, involved in chemokine signaling pathway, cell-cell and cell-extracellular matrix (ECM) adhesion mechanisms. Differentially expressed genes were analyzed, according to predefined pathways and annotated by KEGG (the Kyoto Encyclopedia of Genes and Genomes) [24] and BioCarta (BioCarta Pathways [http://www.biocarta.com/genes/index.asp]) bio-information resources, using the Gene Set Analysis Toolkit. A hypergeometric test was applied in order to determine whether the proportion of the genes falling into each category (down- and up-regulated) was statistically significant. For an over-represented KEGG or Biocarta pathway, a cut-off P value of 0.01was selected. All displayed values were expressed on a logarithm scale, and it should be noted that generally one gene can participate in more than one KEGG or BioCarta pathway.
An analysis of Gene Ontology (GO) was performed on selected genes. The degree of enrichment or depletion of the GO category for each group of genes was calculated, as represented by the specific expression pattern, comparing the enrichment/depletion with that which could have been expected by chance alone, using the two-sided Fisher’s exact test.
RNA isolation and quantitative real-time polymerase chain reaction (RT-PCR)
A quantitative RT-PCR analysis was performed individually on an independent set of new patients in order to validate the microarray-derived results. RNA was extracted from 5 different samples of oophorus cumuli, which had been isolated from patients with endometriosis, and from 7 samples of oophorus cumuli, which had been isolated from control patients; both extractions deployed the commercially available RNAqueous micro kit (Ambion, TX, USA), as described previously. Total RNA was reverse transcribed to cDNA, using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Samples were stored at −20 °C until use. Regarding quantitative TaqMan RT-PCR, master mix and TaqMan gene expression assays for GAPDH (glyceraldehyde 3-phosphate dehydrogenase, Hs99999905_m1) control, TNFAIP6 (Hs01113602_m1), CD44 (Hs01075861_m1), TENASCIN C (Hs01115665_m1), INTEGRIN β2 (Hs00164957_m1), IL-8 (Hs00174103 m1), CXCL2 (Hs00601975_m1), CYP11A1 (Hs00167984_m1), RUNX1 (Hs00231079_m1), NLRP2 (Hs01546932_m1) and ARHGAP10 (Hs00226305_m1) were obtained from Applied Biosystems. Samples were run in duplicate using the Step-One RT-PCR system (Applied Biosystems, Foster City, CA, USA). Relative changes in gene expression between patients with endometriosis and controls were determined using the ΔΔCt method. Levels of the target transcript were normalized to a GAPDH endogenous control, constantly expressed in both groups (ΔCt). Additional subtractions were performed for ΔΔCt values between untreated and treated samples ΔCtvalues. Final values were expressed as a fold of induction (FOI).
Statistical analysis
The background subtraction and normalization of probe set intensities was performed for the statistical analysis using the Robust Multiarray Analysis (RMA) method, as described by Irizarry et al. [25]. Gene expression intensity was compared in order to identify differentially expressed genes, using a moderated t test and a Bayes smoothing approach, which had been developed for a low number of replicates [26]. To compensate for the effect of multiple testing, the false discovery rate was estimated from p values, as derived from moderated t test statistics [27]. The analysis was performed using the affylmGUI Graphical User Interface for the limma microarray package (Bioconductor Software) [28]. A comparison between patient and cycle characteristics was performed by deploying the adequate non parametric test (Mann–Whitney U-test).
Results
Gene expression profiling in CC of patients with severe endometriosis
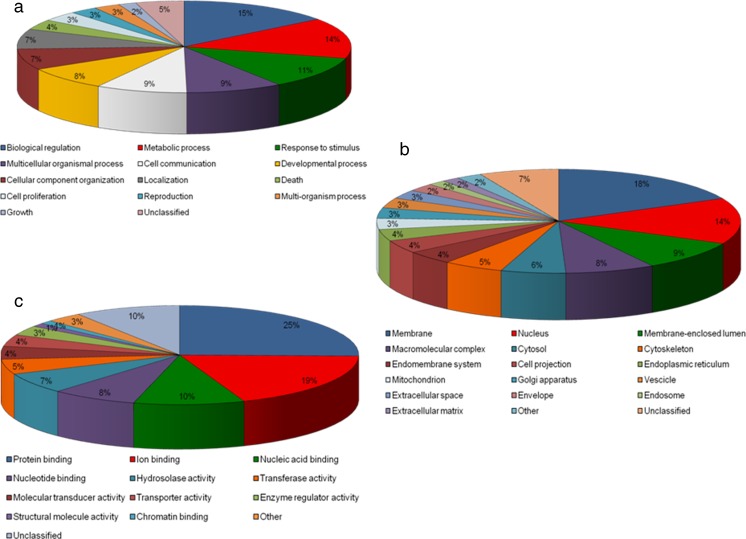
A microarray analysis was performed in order to characterize the differential gene expression profiles in CC of patients with severe endometriosis, using an Affymetrix platform on pooled RNA samples from patients with endometriosis and control patients. This analysis permitted us to obtain a list of differentially expressed genes (DEGs) in the CC of women with severe endometriosis, as compared to healthy women. Specifically, this list was screened by considering genes with p < 0.05 as only significant and fold change ≥1.5 (linear scale). Thus, a list of 595 DEGs between the two analysed groups (320 down-regulated and 275 up-regulated) (Table S1) was obtained. The 595 DEGs were further analyzed by means of the Gene Set Analysis Toolkit software and distributed among the three different GO categories (biological process, molecular function and cell component), making use of pie charts (Fig. 1). A GO analysis indicated that most DEGs fell into the following main sub-categories: the biological process category, biological regulation, metabolic process, response to stimulus and cell communication; the molecular function category: protein, ion, nucleic acid and nucleotide binding; and the cell component category membrane: nucleus, membrane-enclosed lumen and macromolecular complex. Furthermore, it was observed that a high percentage of these genes was shared by the following sub-categories: membrane, extracellular space, extracellular matrix, cell projection and cell communication.
Fig. 1.
Pie charts of GO categories. The diagrams summarize the expression profile microarray data of 595 DEGs, distributed amongst the three different Gene Ontology (GO) categories: (a) Biological Process, (b) Molecular Function and (c) Cell Component. Each category is divided into several sub-categories
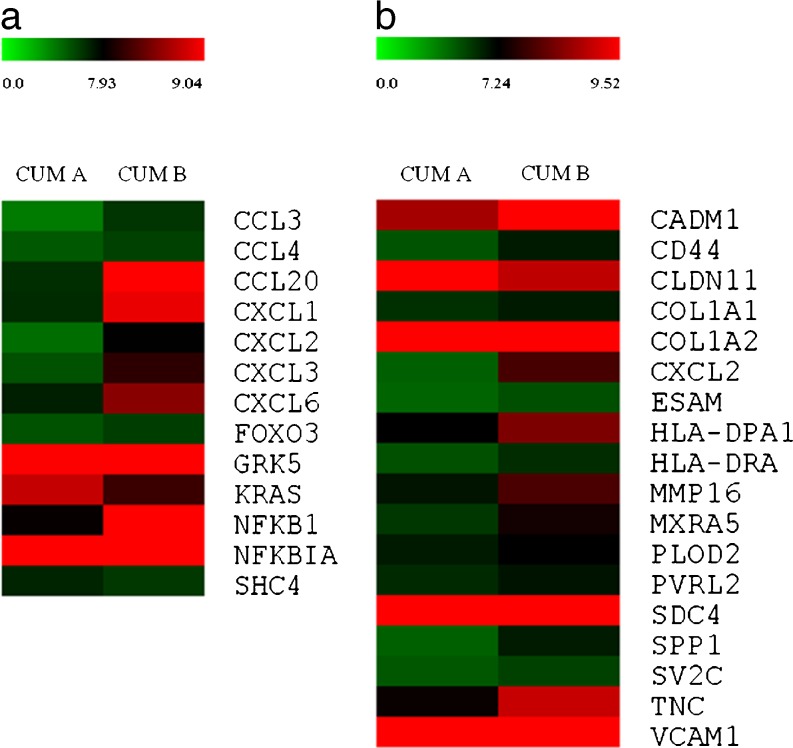
The integrated analysis resulting from different databases (especially, KEGG and the Biocarta bio-information resources) enabled us to identify the main molecular pathways which had been altered in CC in patients with severe endometriosis. This analysis indicated that the majority of genes showing a deregulation in expression levels was included in the following main pathways: cell-cell and cell-ECM interaction (19 genes), chemokine signalling (17 genes), lipid and lipoprotein metabolism (15genes), cytokine-cytokine receptor interaction (14 genes), myometrial relaxation and contraction (12 genes), TNF-alpha/NF-kB signalling (11 genes), NOD-like receptor signalling (9 genes), apoptosis (7 genes), Toll-like receptor signalling (7 genes) and cumulus-oocyte complex formation and maturation (6 genes). However, our attention was focused mainly on the genes involved in cross-talk between oocyte and CC, and the inflammatory response. These two pathways were grouped into two distinct heat maps, using the MultiExperiment Viewer program (MeV v4.8) (Fig. 2). Indeed, the most significant changes in expression levels were observed in key genes involved in the chemokine signalling pathway (Fig. 2a), and cell-cell and cell-ECM adhesion mechanisms (Fig. 2b).
Fig. 2.
Heat maps of DEGs involved in main pathways, deregulated in the CC of patients with endometriosis. The clustering of DEGs involved in (a) chemokine signaling pathway and (b) cell-cell and cell-ECM adhesion mechanisms, identified by integrated analyses using KEGG and BioCarta databases. For an over-represented KEGG or BioCarta pathway, a cut-off p value of 0.01 was selected. The heat maps were generated from microarray data, reflecting gene expression values (p < 0.05 and fold change ≥1.5) in the cumulus oophorus cells of patients with severe endometriosis (Group a) in comparison to control cells (Group b). Each row represents the expression levels for a single gene tested for the two experimental conditions. Each column shows the expression levels for the genes, which were tested for each group of individuals. The absolute expression value (log scale) of each gene has been derived from the mean of two replicates. The color scale bar on the top represents signal intensity variations, ranging from green (poorly expressed genes) to red (highly expressed genes). The black boxes indicate intermediate expression values.
Of interest, almost all genes involved in the chemokine signalling pathway (Fig. 2a), and cell-cell and cell-ECM adhesion mechanisms (Fig. 2b) pathways (except KRAS and SHC4,and CLDN11 and VCAM1 respectively) underwent a negative regulation of the expression in the CC of patients with severe endometriosis. Specifically, the most down-regulated genes belonged to the chemokine group, including CXCL2, which demonstrated the highest value for fold change (−10.3), CCL20 (−8.93), CXCL3 (−6.62) and IL8 (−5.08). Of the DEGs involved in cell-cell and the cell-ECM interaction pathway, significant decreases in expression levels were observed in SPP1 (−3.86), MXRA5 (−3.41), TENASCINC (−3.22), CD44 (−3.01). Finally, the microarray analysis revealed a reduction in the expression levels of genes regulating COC formation and maturation, such as TNFAIP6, INHBA, PTGER4, PTGER2, FSTL3 and EREG.
Real time analysis
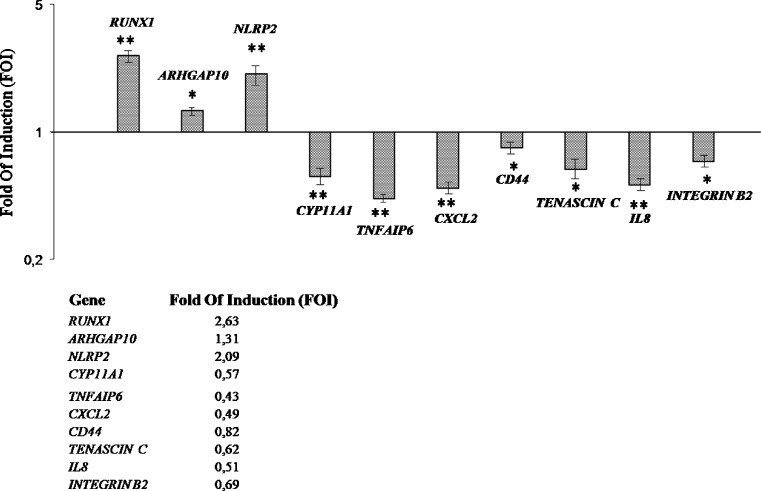
It was decided to analyse the expression levels of mRNA in individual endometriosis samples as compared with controls through single RT-PCR in order to confirm the microarray analysis on pooled cumuli. The expression of the seven down-regulated selected genes (CYP11A1, TNFAIP6, CXCL2, CD44, TENASCINC, IL8 and INTEGRIN Β2) was investigated because they belong to two important biological classes involved in oophorus cumulus and oocyte crosstalk, such as cell-cell and cell-matrix adhesion and the chemokine pathways. Three up-regulated genes (RUNX1, ARHGAP10 and NLRP2) were selected because they participate in the regulatory mechanisms of important physiological processes. The data confirmed the decreased expression of CYP11A1, TNFAIP6, CXCL2, CD44, TENASCINC, IL8 and INTEGRIN Β2 genes in oophorus cumulus cells, which had been collected from endometriosis patients as compared to those collected from controls, and a higher expression of RUNX1, ARHGAP10 and NLRP2 in the same samples (Fig. 3).
Fig. 3.
An RT-PCR analysis of expression levels of selected differentially expressed genes in oophorus cumulus cells from endometriosis patients vs control. It is shown the relative expression of the mRNA of differentially expressed genes in oophorus cumulus cells from endometriosis patients (n = 5), normalized for GAPDH and plotted as a fold of increase over cumulus cells of control patients (n = 7). Results represent the mean ± SD (*P < 0.001; **P < 0.05)
Discussion
The mechanisms of endometriosis-associated infertility have been debated for many years and they are still held to be unclear. Excluding altered pelvic anatomy, these mechanisms include: abnormal folliculogenesis, elevated oxidative stress, altered immune function and reduced endometrial receptivity. In IVF cycles, these factors lead to poor oocyte quality, impaired fertilization and implantation [18]. There is clinical evidence that oocyte competence is the key mechanism impaired in endometriosis-related infertility [7, 29, 30].
The pre-ovulatory oocyte is surrounded by several layers of CC, tightly connected to each other and the oocyte via gap junctions and intercellular membrane processes, thereby establishing bi-directional crosstalk. This interaction is crucial for the functioning of both of the cytotypes, conferring the competence required for fertilization and embryogenesis on the oocyte, through the exchange of nutrients, growth factors, paracrine signals, secondary messengers and other regulatory molecules [31–34]. For these reasons, the analysis of the gene expression profile of CC could provide information regarding oocyte competence. Various microarray-based studies into CC in humans and other mammals have been performed in recent years [9–17, 35, 36].
A transcriptomic analysis of CC derived from endometriosis patients is still lacking. The aim of this study was to compare the transcript profiles of CC isolated from patients with severe endometriosis and controls by means of a wide-genome expression analysis. A further objective of this analysis was to identify any pathway potentially responsible for the low fertilization and implantation rates observed in endometriosis patients. In both groups, the CC associated with mature oocytes of all patients were pooled together. This procedure was mandatory due to the very low quantity of obtained RNA and it has been used by several authors [9, 12, 14, 35, 37–39]. However, the results obtained through conducting microarrays have been validated by using RT-PCR on an individual set of patients. Performing the microarray analysis, 595 genes (320 down-regulated and 275 up-regulated) were identified as having been differentially expressed between those patients suffering from endometriosis and the control patients. The genes chosen for the RT-PCR validation (TNFAIP6, CD44, TENASCIN C, INTEGRIN Β2, IL8, CXCL2,CYP11A1, RUNX1, NLRP2, ARGHAP10) were selected according to their biological relevance in the oocyte maturation process and taking into account the differences in folds of expression. The RT-PCR results confirmed the microarray results for all the analysed genes.
Several functional pathways were identified when the approach outlined in this Paper was adopted. These pathways share many genes, thereby implying that a single pathway cannot be considered as a separate and individualized process. Numerous pathways belong to the field of immunity (Toll-like receptor signalling, NOD-like receptor signalling, chemokine signalling, cytokine-cytokine receptor interaction, TNF-alpha/NF-kB signalling) and cell-cell and cell-ECM adhesion mechanisms, and the vast majority of the genes involved in these pathways are down-regulated in endometriosis CC.
Toll-like and NOD-like receptors are known as Pattern Recognition Receptors (PRR) and are implicated in innate immunity [40]. These receptors recognize pathogen-associated molecular patterns (PAMP), produced by pathogens, and damage-associated molecular patterns (DAMP), which are released by the damaged cells of the human organism. After contact with these molecules, Toll-like and NOD-like receptors activate NFKB, a transcription factor, which induces the production of chemokines and inflammatory cytokines, such as: Tnf alpha, IL1, IL6, IL8, IL12 and IL18.
Differing evidence supports the role of inflammation and the immune response in granulosa and cumulus cells during the ovulation process [41, 42]. Follicular rupture and ovulation are characterized by inflammation-like processes, such as matrix metalloproteinase expression and cytokine production [43, 44]. Specifically, members of the innate immune system, such as the Toll-like receptor, have been identified as expressed/induced in the granulosa/CC of mouse, bovine and human ovaries [41]. The activation of these receptors leads to the activation of many genes, including NFKB and other transcription factors, cytokines (mainly IL6) and chemokines, which may improve the COC expansion. Toll-like receptors may be also involved in fertilization, as it has been demonstrated that sperm is able to induce the release of cytokines and chemokines from CC, thereby enhancing the fertilization process.
Cytokine function in the ovary has also been described as a process promoting follicular growth, steroidogenesis, recruitment and the activation of leukocytes, all of which are necessary for ovulation and tissue remodelling during ovulation [45]. Significant changes in the expression levels of key genes, which are involved in chemokine signaling, were observed and down-regulated in endometriosis CC as compared with the controls. Sarapik and colleagues have reported that various cytokines (such as IL8) in the follicular fluid of infertile women are correlated with a successful pregnancy following IVF treatments [46]. The IL8 in this study appeared to be down-regulated in endometriosis CC as compared with the controls.
CXCL2 is an angiogenic chemokine produced by macrophages, endothelial, epithelial and tumor cells, which has potent chemotactic activity for neutrophils; it thus plays an important role in cytokine-induced inflammatory and immune cell-mediated responses [47]. Little data is present in the literature regarding the role of this chemokine in contributing to oocyte development. Schindler and colleagues have demonstrated that CXCL2 is present in ovarian trascriptome during primordial follicle assembly [48]. CXCL2 appeared to be the most down-regulated gene in this study of endometriosis patients, thus indicating a key role for this chemokine in contributing to tissue remodeling during oocyte maturation.
Several proteins have been identified as being essential for extracellular matrix formation and the stability of COC. Of these, the Tumor necrosis factor alpha-induced protein 6 (TNFAIP6), an inflammation-associated protein, has been widely studied in the formation of the COC extracellular matrix during ovulation [49, 50]. TNFAIP6 proved to be significantly down-regulated in this study of endometriosis CC. Among the other DEGs involved in cell-cell and cell-ECM adhesion, CD44, which encodes for a cell adhesion receptor, appeared to be down-regulated in endometriosis CC. This glycoprotein is present in mature oocytes and preimplantation embryos in various species of mammals, such as humans, but not in immature oocytes [51]. Its expression in the granulosa cells of endometriosis patients was described as significantly depressed, thereby suggesting a correlation with the poor fecundity rate observed in patients with endometriosis [52]. Together with TNFAIP6 and CD44, other molecules involved in cell-cell and cell-ECM interactions (TENASCIN C and INTEGRIN Β2) have been found to be down-regulated in CC from endometriosis patients. Previously, the presence of TENASCIN C has been demonstrated in the literature relating to the cumulus oophorus matrix [53]. Our findings indicate that the down-regulation of these molecules could affect the cell-cell interaction in the COC of endometriosis patients, and this knowledge could be taken into consideration in further studies for an improved understanding of oocyte impairment in patients with endometriosis.
Concerning the steroidogenic pathway, CYP11A encodes for the side chain cleavage P450 enzyme, the latter which converts cholesterol to pregnenolone, thus participating in estrogen synthesis. This gene has been observed to be down-regulated in the CC of patients with endometriosis and it could be another marker of the impaired function of these cells. RUNX1 encodes for a proangiogenic factor and it has been found to be significantly up-regulated in the CC of endometriosis patients. It has already been described as up-regulated in rat endometriotic lesions [54]. Whilst belonging to a family encoding for proteins which are involved in the regulation of inflammatory processes and cell apoptosis, NLRP2 has been found to be up-regulated in endometriosis CC and it has been described in the oocytes and granulosa cells of mice during folliculogenesis [55].
ARHGAP10 (RHOGTPase activating protein 10) has been found to be up-regulated in endometriosis CC. However, there is no experimental evidence of a direct correlation between this gene and endometriosis. In one study regarding gene expression in CC as a prognostic factor for embryo viability, ARHGAP10 was overexpressed in embryos with early cleavage, in comparison with embryos which did not cleave early [14]. The association with other factors involved in angiogenesis was claimed by the Authors of this study as the main reason for this up-regulation.
We believe the data presented in this Paper to be the first evidence of the involvement of these up-regulated genes in the CC of patients with endometriosis but further investigation is required to improve our understanding of the relationship between these molecules and the disease. In conclusion, our data is consistent with the down-regulation in patients with endometriosis of a large number of genes involved in key regulatory pathways. We have reported a gene expression analysis which could offer an improved understanding of the molecular mechanism responsible for the low fertilization and implantation rates in patients with endometriosis. Collectively, these findings appear to be merit further investigation into the possible markers of fertilization failure in patients with severe endometriosis.
Electronic supplementary material
(DOC 30 kb)
Acknowledgments
Declaration of interest
The Authors of this Paper declare there to be no conflict of interest which could be perceived as prejudicing the impartiality of the research reported herein.
Funding
This research did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector; the funds were provided directly by the ANDROS Day Surgery Clinic, Palermo, Italy.
Footnotes
Capsule Several genes involved in the chemokine mediated-signaling pathway and in the cross-talk between cumulus cells and the oocyte are down-regulated in the cumulus cells of patients with endometriosis.
The first two authors contributed equally to this work
References
- 1.Barnhart K, Dunsmoor-Su R, Coutifaris C. Effect of endometriosis on in vitro fertilization. Fertil Steril. 2002;77:1148–55. doi: 10.1016/S0015-0282(02)03112-6. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy S, Bergqvist A, Chapron C, D’Hooghe T, Dunselman G, Greb R, et al. On behalf of the ESHRE special interest group for endometriosis and endometrium guideline development group. ESHRE guideline for the diagnosis and treatment of endometriosis. Hum Reprod. 2005;20:2698–704. doi: 10.1093/humrep/dei135. [DOI] [PubMed] [Google Scholar]
- 3.The Practice Committee of the American Society for Reproductive Medicine Endometriosis and infertility: a committee opinion. Fertil Steril. 2012;98:591–8. doi: 10.1016/j.fertnstert.2012.05.031. [DOI] [PubMed] [Google Scholar]
- 4.Lin X, Wei M, Tong X, Xu W, Zhou F, Huang Q, et al. Outcome of in vitro fertilization in endometriosis-associated infertility: a 5-year database cohort study. Chin Med J. 2012;125:2688–93. [PubMed] [Google Scholar]
- 5.Society for Assisted Reproductive Technology, the American Society for Reproductive Medicine Assisted reproductive technology in the United States: 2010 results generated from the American Society for Reproductive Medicine/Society for Assisted Reproduction registry. Available at: www.sart.org
- 6.Dunselman G, Vermeulen N, Becker C, Calhaz-Jorge C, D'Hooghe T, De Bie B, et al. ESHRE guideline: management of women with endometriosis. Hum Reprod. 2014;29:400–12. doi: 10.1093/humrep/det457. [DOI] [PubMed] [Google Scholar]
- 7.Simón C, Gutierrez A, Vidal A, Tarin JJ, Remohí J, Pellicer A. Outcome of patients with endometriosis in assisted reproduction: results from in-vitro fertilization and oocyte donation. Hum Reprod. 1994;9:725–9. doi: 10.1093/oxfordjournals.humrep.a138578. [DOI] [PubMed] [Google Scholar]
- 8.Garrido N, Navarro J, García-Velasco J, Remohí J, Pellicer A, Simón C. The endometrium versus embryonic quality in endometriosis-related infertility. Hum Reprod Update. 2002;8:95–103. doi: 10.1093/humupd/8.1.95. [DOI] [PubMed] [Google Scholar]
- 9.Adriaenssens T, Wathlet S, Segers I, Verheyen G, De Vos A, Van der Elst J, et al. Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Hum Reprod. 2010;25:1259–70. doi: 10.1093/humrep/deq049. [DOI] [PubMed] [Google Scholar]
- 10.Assou S, Haouzi D, Mahmoud K, Aouacheria A, Guillemin Y, Pantesco V, et al. A non-invasive test for assessing embryo potential by gene expression profiles of human cumulus cells: a proof of concept study. Mol Hum Reprod. 2008;14:711–9. doi: 10.1093/molehr/gan067. [DOI] [PubMed] [Google Scholar]
- 11.Feuerstein P, Cadoret V, Dalbies-Tran R, Guerif F, Bidault R, Royere D. Gene expression in human cumulus cells: one approach to oocyte competence. Hum Reprod. 2007;22:3069–77. doi: 10.1093/humrep/dem336. [DOI] [PubMed] [Google Scholar]
- 12.Hamel M, Dufort I, Robert C, Gravel C, Leveille MC, Leader A, et al. Identification of differentially expressed markers in human follicular cells associated with competent oocytes. Hum Reprod. 2008;23:1118–27. doi: 10.1093/humrep/den048. [DOI] [PubMed] [Google Scholar]
- 13.McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, et al. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 2004;19:2869–74. doi: 10.1093/humrep/deh535. [DOI] [PubMed] [Google Scholar]
- 14.van Montfoort AP, Geraedts JP, Dumoulin JC, Stassen AP, Evers JL, Ayoubi TA. Differential gene expression in cumulus cells as a prognostic indicator of embryo viability: a microarray analysis. Mol Hum Reprod. 2008;14:157–68. doi: 10.1093/molehr/gam088. [DOI] [PubMed] [Google Scholar]
- 15.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke W, et al. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod. 2011;26:1035–51. doi: 10.1093/humrep/der036. [DOI] [PubMed] [Google Scholar]
- 16.Brannian J, Eyster K, Mueller BA, Bietz MG, Hansen K. Differential gene expression in human granulosa cells from recombinant FSH versus human menopausal gonadotropin ovarian stimulation protocols. Reprod Biol Endocrinol. 2010;8:25. doi: 10.1186/1477-7827-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grondahl ML, Borup R, Lee YB, Myrhoj V, Meinertz H, Sorensen S. Differences in gene expression of granulosa cells from women undergoing controlled ovarian hyperstimulation with either recombinant follicle-stimulating hormone or highly purified human menopausal gonadotropin. Fertil Steril. 2009;91:1820–30. doi: 10.1016/j.fertnstert.2008.02.137. [DOI] [PubMed] [Google Scholar]
- 18.Saito H, Seino T, Kaneko T, Nakahara K, Toya M, Kurachi H. Endometriosis and oocyte quality. Gynecol Obstet Invest. 2002;53(Suppl 1):46–51. doi: 10.1159/000049424. [DOI] [PubMed] [Google Scholar]
- 19.Sifer C, Benifla JL, Bringuier AF, Porcher R, Blanc-Layrac G, Madelenat P, et al. Could induced apoptosis of human granulosa cells predict in vitro fertilization-embryo transfer outcome? A preliminary study of 25 women. Eur J Obstet Gynecol Reprod Biol. 2002;103:150–3. doi: 10.1016/S0301-2115(02)00043-X. [DOI] [PubMed] [Google Scholar]
- 20.American Society for Reproductive Medicine Revised american society for reproductive medicine classification of endometriosis: 1996. Fertil Steril. 1997;67:817–21. doi: 10.1016/S0015-0282(97)81391-X. [DOI] [PubMed] [Google Scholar]
- 21.Volpes A, Sammartano F, Coffaro F, Mistretta V, Scaglione P, Allegra A. Number of good quality embryos on day 3 is predictive for both pregnancy and implantation rates in in vitro fertilization/intracytoplasmic sperm injection cycles. Fertil Steril. 2004;82:1330–6. doi: 10.1016/j.fertnstert.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 22.Patrat C, Kaffel A, Delaroche L, Guibert J, Jouannet P, Epelboin S, et al. Optimal timing for oocyte denudation and intracytoplasmic sperm injection. Obstet Gynecol Int. 2012;2012:403531. doi: 10.1155/2012/403531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004; 3:Article3 [DOI] [PubMed]
- 27.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 28.Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–9. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- 29.Pellicer A, Albert C, Mercader A, Bonilla-Musoles F, Remohi J, Simón C. The follicular and endocrine environment in women with endometriosis: local and systemic cytokine production. Fertil Steril. 1998;70:425–31. doi: 10.1016/S0015-0282(98)00204-0. [DOI] [PubMed] [Google Scholar]
- 30.Yanushpolsky EH, Best CL, Jackson KV, Clarke RN, Barbieri RL, Hornstein MD. Effects of endometriomas on oocyte quality, embryo quality, and pregnancy rates in in vitro fertilization cycles: a prospective, case-controlled study. J Assist Reprod Genet. 1998;15:193–7. doi: 10.1023/A:1023048318719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buccione R, Schroeder AC, Eppig JJ. Interactions between somatic cells and germ cells throughout mammalian oogenesis. Biol Reprod. 1990;43:543–7. doi: 10.1095/biolreprod43.4.543. [DOI] [PubMed] [Google Scholar]
- 32.Motta PM, Makabe S, Naguro T, Correr S. Oocyte follicle cells association during development of human ovarian follicle. A study by high resolution scanning and transmission electron microscopy Arch Histol Cytol. 1994;57:369–94. doi: 10.1679/aohc.57.369. [DOI] [PubMed] [Google Scholar]
- 33.Albertini DF, Combelles CM, Benecchi E, Carabatsos MJ. Cellular basis for paracrine regulation of ovarian follicle development. Reproduction. 2001;121:647–53. doi: 10.1530/rep.0.1210647. [DOI] [PubMed] [Google Scholar]
- 34.Assidi M, Dufort I, Ali A, Hamel M, Algriany O, Dielemann S, et al. Identification of potential markers of oocyte competence expressed in bovine cumulus cells matured with follicle-stimulating hormone and/or phorbol myristate acetate in vitro. Biol Reprod. 2008;79:209–22. doi: 10.1095/biolreprod.108.067686. [DOI] [PubMed] [Google Scholar]
- 35.Assou S, Anahory T, Pantesco V, Le Carrour T, Pellestor F, Klein B, et al. The human cumulus–oocyte complex gene-expression profile. Hum Reprod. 2006;21:1705–19. doi: 10.1093/humrep/del065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.May-Panloup P, Ferre-L’Hotellier V, Moriniere C, Marcaillou C, Lemerle S, Malinge MC, et al. Molecular characterization of corona radiata cells from patients with diminished ovarian reserve using microarray and microfluidic-based gene expression profiling. Hum Reprod. 2012;27:829–43. doi: 10.1093/humrep/der431. [DOI] [PubMed] [Google Scholar]
- 37.Bettegowda A, Patel OV, Lee KB, Park KE, Salem M, Yao J, et al. Identification of novel bovine cumulus cell molecular markers predictive of oocyte competence: functional and diagnostic implications. Biol Reprod. 2008;79:301–9. doi: 10.1095/biolreprod.107.067223. [DOI] [PubMed] [Google Scholar]
- 38.Assidi M, Montag M, Van Der Ven K, Sirard MA. Biomarkers of human oocyte developmental competence expressed in cumulus cells before ICSI: a preliminary study. J Assist Reprod Genet. 2011;28:173–88. doi: 10.1007/s10815-010-9491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatta V, Tatone C, Ciriminna R, Vento M, Franchi S, Aurora M, et al. Gene expression profiles of cumulus cells obtained from women treated with recombinant human luteinizing hormone + recombinant human follicle-stimulating hormone or highly purified human menopausal gonadotropin versus recombinant human follicle-stimulating hormone alone. Fertil Steril. 2013;99:2000–8. doi: 10.1016/j.fertnstert.2013.01.150. [DOI] [PubMed] [Google Scholar]
- 40.Fritz JH, Girardin SE. How Toll-like receptors and Nod-like receptors contribute to innate immunity in mammals. J Endotoxin Res. 2005;11:390–4. doi: 10.1179/096805105X76850. [DOI] [PubMed] [Google Scholar]
- 41.Liu Z, Shimada M, Richards JS. The involvement of the Toll-like receptor family in ovulation. J Assist Reprod Genet. 2008;25:223–8. doi: 10.1007/s10815-008-9219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, de Matos DG, Fan HY, Shimada M, Palmer S, Richards JS. Interleukin-6: an autocrine regulator of the mouse cumulus cell-oocyte complex expansion process. Endocrinology. 2009;150:3360–8. doi: 10.1210/en.2008-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards JS, Russell DL, Robker RL, Dajee M, Alliston TN. Molecular mechanisms of ovulation and luteinization. Mol Cell Endocrinol. 1998;145:47–54. doi: 10.1016/S0303-7207(98)00168-3. [DOI] [PubMed] [Google Scholar]
- 44.Gerard N, Caillaud M, Martoriati A, Goudet G, Lalmanach AC. The interleukin-1 system and female reproduction. J Endocrinol. 2004;180:203–12. doi: 10.1677/joe.0.1800203. [DOI] [PubMed] [Google Scholar]
- 45.Buscher U, Chen FC, Kentenich H, Schmiady H. Cytokines in the follicular fluid of stimulated and non-stimulated human ovaries; is ovulation a suppressed inflammatory reaction? Hum Reprod. 1999;14:162–6. doi: 10.1093/humrep/14.1.162. [DOI] [PubMed] [Google Scholar]
- 46.Sarapik A, Velthut A, Haller-Kikkatalo K, Faure GC, Bene MC, de Carvalho Bittencourt M, et al. Follicular proinflammatory cytokines and chemokines as markers of IVF success. Clin Dev Immunol. 2012;2012:606459. doi: 10.1155/2012/606459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehrad B, Keane MP, Strieter RM. Chemokines as mediators of angiogenesis. Thromb Haemost. 2007;97:755–62. [PMC free article] [PubMed] [Google Scholar]
- 48.Schindler R, Nilsson E, Skinner MK. Induction of ovarian primordial follicle assembly by connective tissue growth factor CTGF. PLoS One. 2010;5:e12979. doi: 10.1371/journal.pone.0012979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fulop C, Szanto S, Mukhopadhyay D, Bardos T, Kamath RV, Rugg MS, et al. Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development. 2003;130:2253–61. doi: 10.1242/dev.00422. [DOI] [PubMed] [Google Scholar]
- 50.Milner CM, Day AJ. TSG-6: a multifunctional protein associated with inflammation. JCell Sci. 2003;116:1863–73. doi: 10.1242/jcs.00407. [DOI] [PubMed] [Google Scholar]
- 51.Campbell S, Swann HR, Aplin JD, Seif MW, Kimber SJ, Elstein M. CD44 is expressed throughout pre-implantation human embryo development. Hum Reprod. 1995;10:425–30. doi: 10.1093/HUMREP/10.6.1571. [DOI] [PubMed] [Google Scholar]
- 52.Ohta N, Saito H, Kuzumaki T, Takahashi T, Ito MM, Saito T, et al. Expression of CD44 in human cumulus and mural granulosa cells of individual patients in in-vitro fertilization programmes. Mol Hum Reprod. 1999;5:22–8. doi: 10.1093/molehr/5.1.22. [DOI] [PubMed] [Google Scholar]
- 53.Familiari G, Verlengia C, Nottola SA, Renda T, Micara G, Aragona C, et al. Heterogeneous distribution of fibronectin, tenascin-C, and laminin immunoreactive material in the cumulus-corona cells surrounding mature human oocytes from IVF-ET protocols–evidence that they are composed of different subpopulations: an immunohistochemical study using scanning confocal laser and fluorescence microscopy. Mol Reprod Dev. 1996;43:392–402. doi: 10.1002/(SICI)1098-2795(199603)43:3<392::AID-MRD14>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 54.Konno R, Fujiwara H, Netsu S, Odagiri K, Shimane M, Nomura H, et al. Gene expression profiling of the rat endometriosis model. Am J Reprod Immunol. 2007;58:330–43. doi: 10.1111/j.1600-0897.2007.00507.x. [DOI] [PubMed] [Google Scholar]
- 55.Peng H, Chang B, Lu C, Su J, Wu Y, Lv P, et al. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLoS One. 2012;7:e30344. doi: 10.1371/journal.pone.0030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 30 kb)