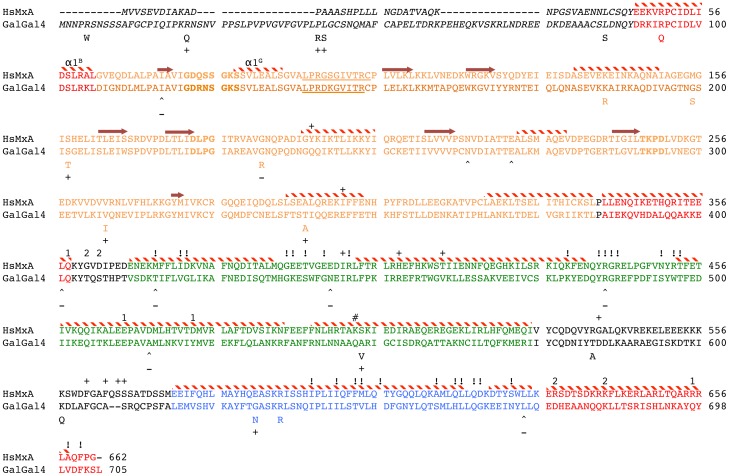
Figure 3. Amino acid alignment of the HsMxA and galGal4.
The different Mx functional domains are represented in colored text (BSE = red, G-domain = orange, Middle stalk domain = green, GTPase effector domain = blue) with the N-terminal region not represented in shown in italics, and the loop regions that connect BSE2 to MD (L1BS) and MD to GED (L4S) in plain text. Positions in orange bold represent the conserved GTPase enzymatic domain and underlined orange text denotes the GTP binding site. Secondary structural elements as described by Gao et al. [23] et al. are indicated above the HsMxA sequence. Alpha helices are represented by red and with stripped bars. Beta strands are represented by arrows. Amino acids with similar numbers above the alignment indicate positions described to interact during oligimerization. HsMxA positions labeled with “#” indicates amino acid with forms hydrogen bonds with the backbone of the α3B, and “!” denote additional positions described to be involved in oligemerization. “+” above the hsMxA indicates amino acid described by Mitchell et al. [30] as under diversifying selection among primate MxA sequences. Amino acid positions in the chMx associated with dNS changes are reflected by the alternate amino acid under the chMx sequence. Positions associated with dS nt changes are indicated by “∧”. chMx positions with evidence of diversifying (dNS) or purifying (dS) selection are indicated with “+” or “−” under the chMx sequence.