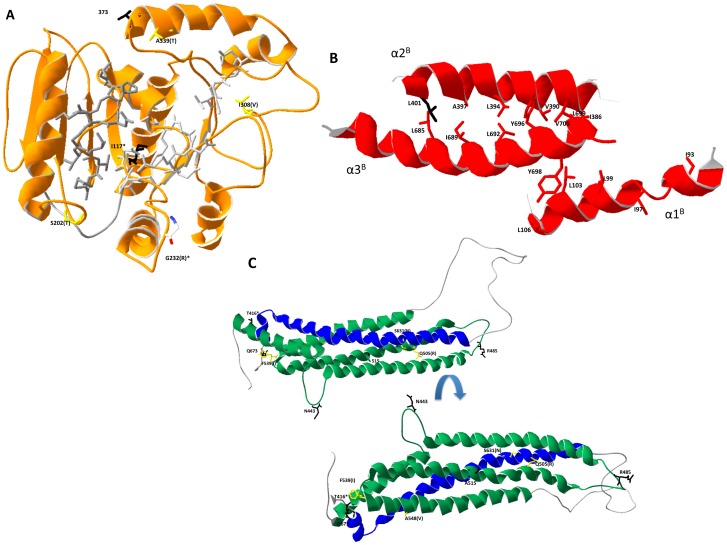
Figure 4. Ribbon structure of the three main functional domains of the Mx protein.
(A) Ribbon structure of the predicted chMx G-domain. The amino acids associated with the GTPase active site are shown in light gray and those associated with GTP binding in dark gray. The amino acids found to be under selection and/or associated with traits are depicted in black (dS) or yellow (dNS). Amino acid 232 had 2 SNP associated with it, one dS and one dNS and is shown in red white and blue. The amino acid and position numbers are next to each selected site with the alternate amino acid indicated in parentheses if applicable. Positions that were associated with both traits and selection are denoted with “*”. (B) Ribbon structure of the interacting BSE elements based on the predicted structure. Position 401, with evidence of purifying selection, is shown in black along with the conserved hydrophobic residues associated with the Lucien zipper (red) interactions. (C) Ribbon structure of the predicted stalk domain. The amino acids found to be under selection and/or associated with traits are depicted in black (dS) or yellow (dNS). The structure was rotated 180° (top vs bottom) in order to be able to visualize all the affected position in the stalk.