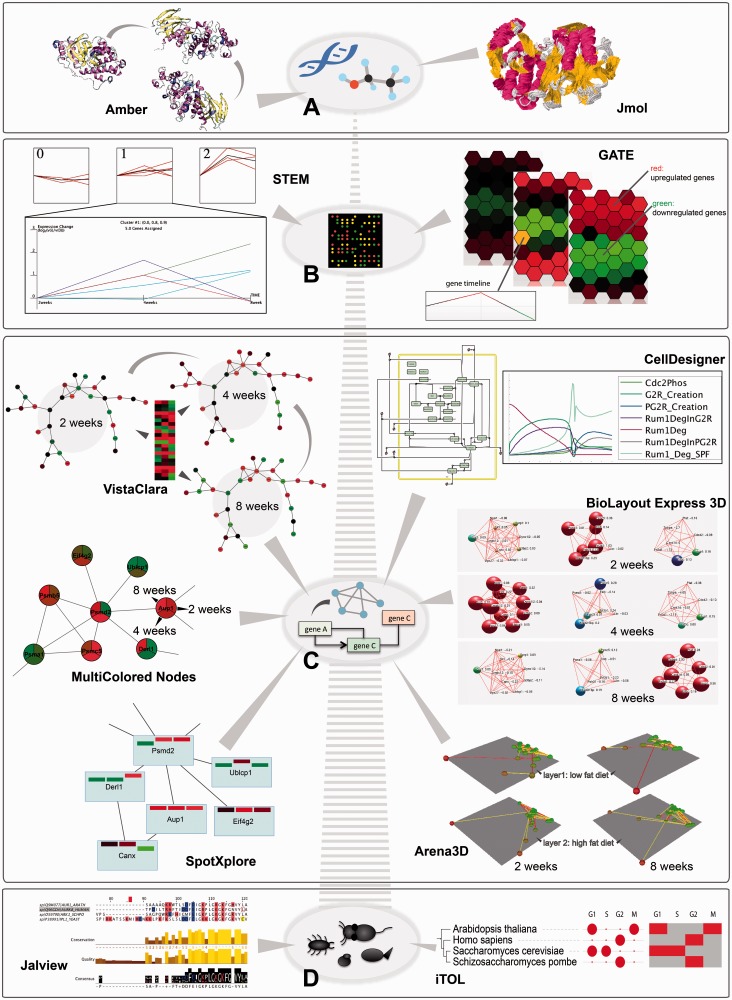
Figure 3:
Temporal depictions of biological processes at different scales are shown, along with a selection of tools that perform the task. (A) At the molecular level, simulations of molecular movement can be followed in an animation using Amber or as trajectory traces using Jmol (example shows MAP kinase P38, as taken from MoDEL library [50]); (B) At the gene level, time course expression data reflecting high fat diet effects on small intestine in mouse (dataset GDS3357 from Gene Expression Ominbus [51]) is visualized in clustered timeline plots using STEM or in an adjacent hexagon display using GATE; (C) At the network level, time course changes can be tracked using different Cytoscape plugins, e.g. by animating colour changes in the network with VistaClara [52], drawing pie chart slices with MultiColored Nodes [53], or using bar charts embedded in the network nodes with SpotXplore [54]. Fluxes through pathways can be simulated deterministically or stochastically and illustrated in line plots using CellDesigner. BioLayout Express 3D simulates changes in gene expression in 3D through colour and node size increase or decrease in an animation (connections represent correlations). Arena3D depicts changes at every time point through colour and clustering on separate 3D layers, corresponding to different phenotypes (low and high fat effects) that can be compared (connections represent correlations). The data used for these examples are the same as in (B). (D) At the organismal level, multiple sequence alignment visualizers, like Jalview, and phylogenetic tree builders, like iTOL, depict evolutionary distances between entities of different organisms. The example shows such depictions for aurora kinase B orthologs in four species. In the case of iTOL, additional time course data can be visualized in the form of discs, heat maps or animations (here we show the phases in the cell cycle where this gene has a periodic peak of transcription, as obtained from Cyclebase [55]).