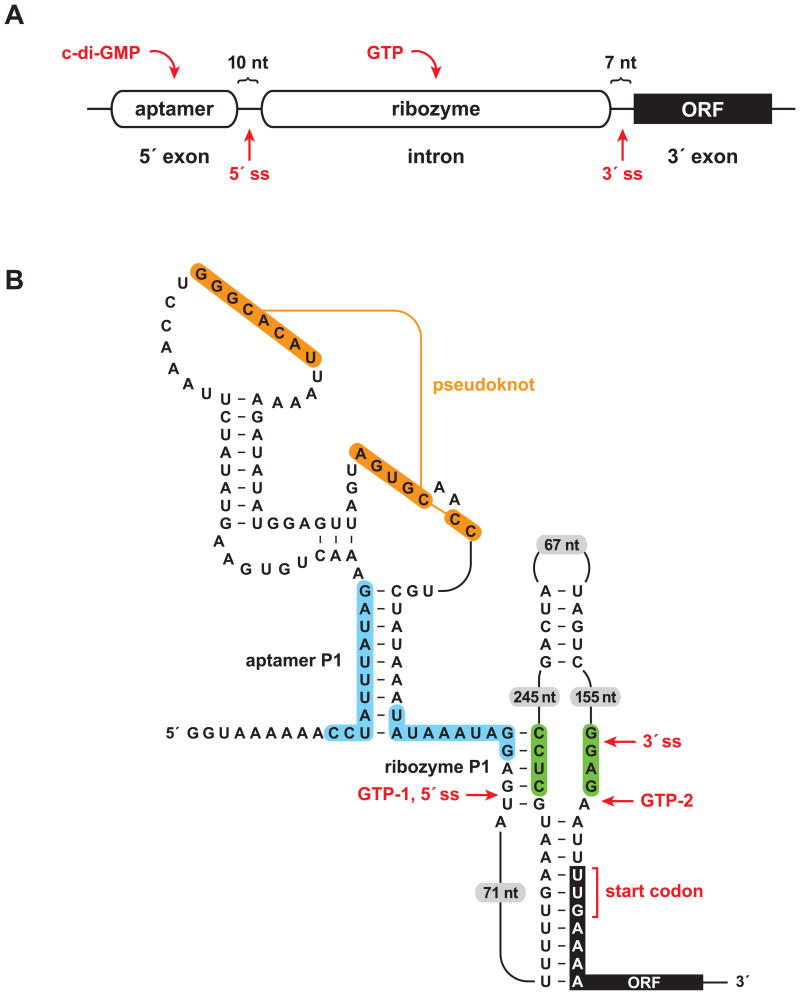
Figure 1.
A sophisticated RNA device. (A) Arrangement of aptamer and ribozyme domains within a naturally-occurring allosteric self-splicing group I ribozyme. The aptamer senses the bacterial second messenger c-di-GMP and the ribozyme requires guanosine or one of its phosphorylated derivatives (e.g., GTP) as a substrate to initiate the first step of splicing. ORF, open reading frame; ss, splice site. (B) Key sequence and secondary structural elements within the allosteric switch. Binding of c-di-GMP to the aptamer domain stabilizes the aptamer P1 stem, which permits formation of the ribozyme P1stem, thereby enabling splicing initiated by GTP attack at the 5′ splice site (GTP-1). This configuration allows translation of the downstream ORF. In the absence of c-di-GMP, alternative pairing (blue shading) precludes formation of the ribozyme P1 stem and allows formation of an alternative ribozyme stem (green shading). This promotes GTP to attack at a position far downstream from the normal 5′ splice site (GTP-2), thus preventing translation of the downstream ORF. Figure adapted from Lee et al., 2010.