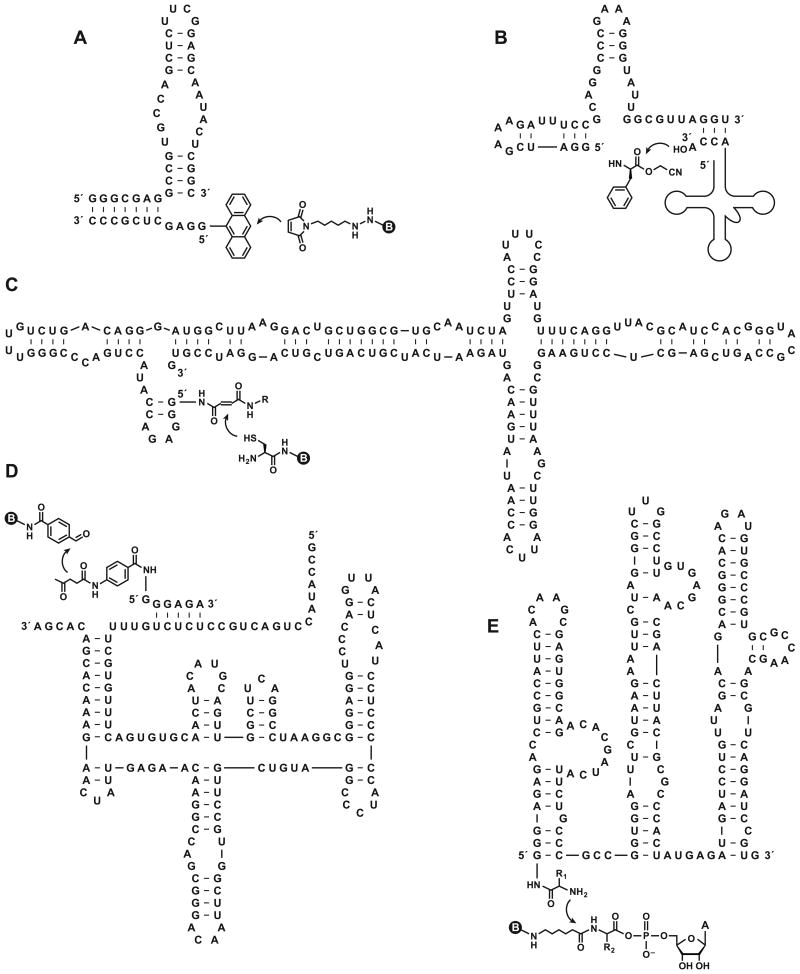
Figure 2.
What do these RNAs have in common? All are in vitro evolved ribozymes that catalyze biologically relevant chemical transformations and all were published in Chemistry & Biology. (A) The “39M38tr” ribozyme, which catalyzes Diels-Alder cycloaddition between biotin maleamide and anthracene that is tethered to the 5′ end of the ribozyme via an alkyl linker (Seelig and Jäschke, 1999). (B) The “Fx3 (Flexizyme)” ribozyme, which catalyzes 3′-aminoacylation of tRNA using the cyanomethyl ester of phenylalanine or other amino acids (Murakami et al., 2003). (C) The “UV5” ribozyme, which catalyzes Michael addition between biotin cysteine and fumaramide that is tethered to the 5′ end of the ribozyme (Sengle et al., 2001). (D) The “11D2” ribozyme, which catalyzes aldol condensation between biotin-linked benzaldehyde-4-carboxamide and levulinic amide that is tethered to the 5′ end of a separate oligonucleotide (Fusz et al., 2005). (E) The “R180” ribozyme, which catalyzes peptide bond formation between an aminoacyl 5′-adenylate and an amino acid that is tethered to the 5′ end of the ribozyme via a disulfide linkage (Zhang and Cech, 1998; Sun et al., 2002). Curved arrow indicates the site of reaction. Circled B indicates a biotin moiety.