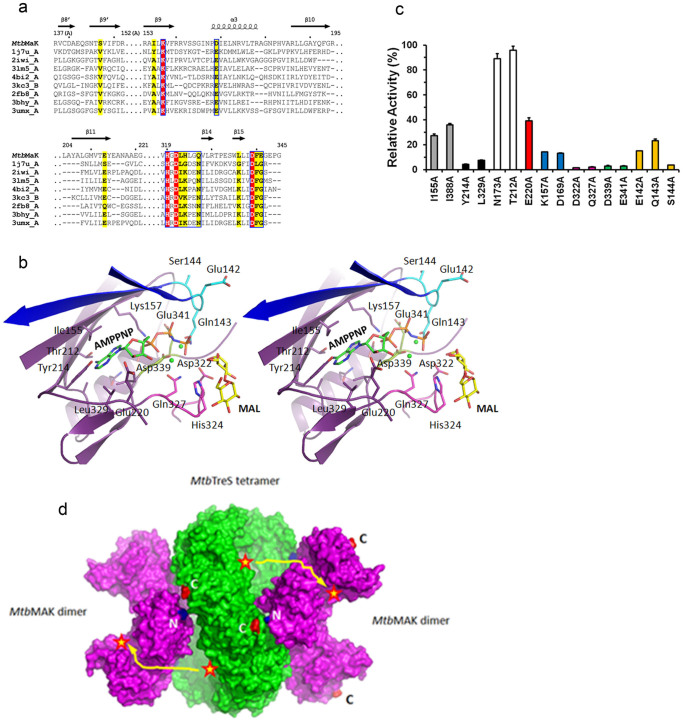
Figure 4. Nucleotide binding site of MtbMaK.
(a). Sequence alignment of nucleotide binding site (NBS) of MtbMaK and its homologues. SSE of MtbMaK are labeled on top of the aligned sequences. Identical residues are highlighted in red, and other conserved residues are highlighted in yellow. (b). Stereo view of AMPPNP-Mg (green sticks and spheres, PDB code 1J7U) superimposed on the NBS of MtbMaK. The C atoms of ‘142EQS144’ loop, HGD motif and DFE motif are colored in cyan, magenta and light green, respectively. Strands β8′ and β9′ from the other subunit of MtbMaK are colored in blue. The side chains of residues potentially interacting with AMPPNP-Mg are shown as sticks. The O and N atoms are colored in red and blue. (c). Mutagenesis of residues from the NBS. A bar graph of relative activity (%) of mutants compared to the wild type enzyme is shown. Error bars represent s.d. (n = 3). (d). Model of hetero-octameric complex of TreS with MaK. Potential interaction of a tetramer of TreS (green color, PDB code 4LXF) with two dimers of MtbMaK (purple color) is shown. Active sites are marked with a red star; putative path of product marked in yellow.