Table 5.
Summary of motifs detected in an unaligned dataset of 199 sequences representative of the diversity of core NBS in Triticeae using MEME. Residues in the Arabidopsis motifs that are identical to the Triticeae ones are indicated in red.
| Motif | Logo | Annotation/position | Significance (e-value and percentage of occurrences) | Homology (if any) to Arabidopsis thaliana CNL motifs |
|---|---|---|---|---|
| 1 |

|
P-loop | 7.7e-1829 (100%) | Triticeae:
G[LV]
GKTTLA[QR]x[VI]YNDx A. thaliana CNL: VGIYGMG GVGKTTIARALF |
| 2 |

|
Kinase 2 | 1.4e-1803 (100%) | Triticeae: L[QK][GD]KR[YF][
LF][I
LV]
V[
LI]
DD[VI]WD A. thaliana CNL: KRF LLVLDDW |
| 3 |

|
Kinase 3a (RNBS-B) | 5.6e-1431 (100%) | Triticeae: x
GSR[I
V][I
L]V
TTRIxDVA A. thaliana CNL: N GCK VLF TTRSEEVC |
| 4 |

|
GLPLA | 8.2e-1120 (100%) | Triticeae: [KE]I[VAL]KK[CL][GK]
G[
LS]
PL A. thaliana CNL: EVAKKCG GLPLALKVI |
| 5 |
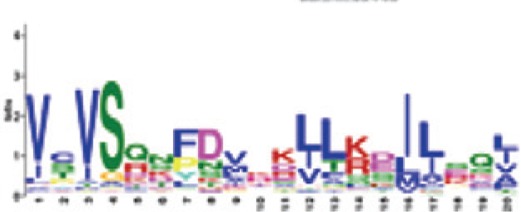
|
RNBS-A | 8.5e-1072 (100%) | Triticeae: VCVSQN[FP]DVxK[LI]L[KR][DE]I[LI][ES]Q[LI] A. thaliana CNL: VKxGFDIVIWVVVSQEFTLKKIQQDILEK |
| 6 |

|
RNBS-C | 3.5e-624 (100%) | Triticeae: DS
WELFx
KR[AI
V]F A. thaliana CNL: KVECLTPEEA WELFQR KV |
| 7 |

|
Between P-loop and RNBS-A | 3.6e-406 (100%) | Triticeae: GHFDCRA[WF] A. thaliana CNL: - |
| 8 |

|
Between Kinase3a and RNBS-C | 1.2e-091 (100%) | Triticeae: [YD]Q[LM]KPL A. thaliana CNL - |
| 9 |

|
Between RNBS-C and GLPLA | 1.6e+075 (100%) | Triticeae: E[LF]EE[IV][GSA] A. thaliana CNL: - |
| 10 |

|
Between Kinase2 and Kinase3a | 4.6e+082 (100%) | Triticeae: W[ED]x[LI]Kx A. thaliana CNL: - |
