Abstract
Background
Highly sensitive markers are urgently needed for the diagnosis and grading of gastric cancer and for managing drug resistance. The recent identification of long-non-coding RNAs (lncRNAs) has provided new approaches for resolving this challenge. The aim of this study was to screen and identify new biomarkers for human gastric cancer from lncRNAs.
Methods
First, we used lncRNA microarrays to conduct a preliminary screening for candidate lncRNAs of gastric cancer biomarkers in both human gastric cancer tissues and in two gastric cancer cell lines, SGC7901 cells and paclitaxel-resistant SGC7901 cells. The lncRNA plasma-cytoma variant translocation 1 (PVT1) was found to exhibit higher expression in both gastric cancer tissues and the SGC7901 paclitaxel-resistant cell line. Quantitative polymerase chain reaction was used for large-scale analysis in a large number of human gastric cancer tissues to verify the involvement of PVT1 in development of gastric cancer. The relationships between PVT1 expression and clinical features were also analyzed.
Results
PVT1 showed higher expression in human gastric cancer tissues than in adjacent non-cancerous tissues and in SGC7901 paclitaxel-resistant cells compared with SGC7901 cells. PVT1 expression was correlated with lymph node invasion of gastric cancer.
Conclusion
PVT1 is a new biomarker for human gastric cancer and may indicate lymph node invasion. Therefore, PVT1 shows potential as a novel therapeutic target for the treatment of gastric cancer and enhancement of paclitaxel sensitivity.
Keywords: microarray analysis, quantitative polymerase chain reaction, lymph node invasion, tumor biomarkers, paclitaxel resistance
Introduction
Gastric cancer remains a major public health concern as it represents the fourth most common cancer and the second leading cause of cancer-related deaths worldwide.1–3 Nevertheless, the incidence and mortality associated with gastric cancer have decreased considerably over the past 50 years in most areas of the world.1 Although conventional diagnostic methods such as gastric endoscopy have contributed to this reduction by enabling early diagnosis, many patients are diagnosed with advanced gastric cancer and have a poor prognosis. Thus, more sensitive tumor markers are urgently needed to improve screening, diagnosis, prognostic evaluation, and tumor grading of gastric cancer. Several potential approaches have been proposed to find suitable marker candidates, and the recent discovery of long-non-coding RNAs (lncRNAs) provides a new opportunity to resolve this challenge. LncRNAs are non-protein-coding transcripts longer than 200 nucleotides4 and are widely distributed in the genome; indeed, more than 90% of the human genome is composed of non-coding RNAs.5 There is mounting evidence that many lncRNAs are dysregulated in various cancers and play important roles in tumorigenesis and invasion.6–8 Thus, lncRNAs may serve as useful tumor markers and therapeutic targets for controlling malignant tumors. In the current study, we investigated the expression of plasmacytoma variant translocation 1 (PVT1), a documented oncogenic lncRNA,9 in gastric cancer and examined its association with clinical features in patients with gastric cancer to reveal its potential as a biomarker and therapeutic target.
Materials and methods
Thirty-three tumor tissues and corresponding non-cancerous tissues were obtained at the time of surgery from June 2012 to October 2013 at the First Affiliated Hospital and Union Hospital of Fujian Medical University, Fuzhou, Fujian, People’s Republic of China. Following excision, the gastric cancer and non-cancerous tissues were immediately frozen in liquid nitrogen and stored at −80°C until use.
The SGC7901 gastric cancer cell line was obtained from the China Center for Type Culture Collection (Wuhan, People’s Republic of China). Paclitaxel-resistant SGC7901 cells were developed in our laboratory and cultured in 450 μg/L paclitaxel. Total mRNA was isolated using TRIzol® Reagent (Life Technologies Corporation, Carlsbad, CA, USA), according to the manufacturer’s protocol.
The paclitaxel-resistant cell line was developed by culture of an SGC7901 gastric cancer cell line in vitro with an increasing gradient of concentrations of paclitaxel intermittently over 6 months. The initial concentration of paclitaxel was 25 μg/L. Forty-eight hours later, medium containing 25 μg/L paclitaxel was replaced with routine medium until the SGC7901 cells were growing in exponential period, after which the paclitaxel concentration was increased to 50 μg/L. Six months later, the final concentration of paclitaxel was 450 μg/L, and the resistance in paclitaxel-resistant SGC7901 cells was detected with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) and compared with normal SGC7901 cells. The MTT assay was performed after 48 hours of incubation with paclitaxel (five concentrations in a multiple proportion dilution from 160 to 10 μg/L), the plates were centrifuged to pellet the cells, the supernatant was removed, and 20 μL of 5 mg/mL MTT was added in 180 μL of medium, followed by incubation for 4 hours at 37°C in a humid, 5% CO2 atmosphere. Next, the plates were centrifuged again, the supernatant was removed, and the insoluble formazan crystals were dissolved in dimethyl sulfoxide. Absorbance was read at 490 nm. The half maximal inhibitory concentration (IC50) was calculated using the Spearman–Kärber cytopathic effect method.10 Before lncRNA microarray analysis, the MTT assay showed IC50 values for SGC7901 and paclitaxel-resistant SGC7901 of 22.16±0.31 μg/L and 141.31±0.9 5 μg/L, respectively. The difference was statistically significant (P<0.001). The drug resistance index for paclitaxel-resistant SGC7901 cells was 6.38.
Microarray analysis
Tissue samples (two gastric cancer tissues and two corresponding non-cancerous tissues) and SGC7901 cells were used to synthesize double-stranded complementary DNA, which was labeled according to the protocol of the Amplification and Labeling Kit for Microarray (CapitalBio Corporation, Beijing, People’s Republic of China). The labeled complementary DNA was hybridized to the Agilent Human LncRNA 4×180 K Expression Microarray (Agilent Technologies Inc., Santa Clara, CA, USA), which contains 37,000 human lncRNAs and 34,000 mRNA probes (CapitalBio Corporation). After hybridization and washing, the processed slides were scanned with a G2565CA microarray scanner (Agilent Technologies). Raw data were extracted as paired files using Feature Extraction software (Agilent Technologies). The raw data were normalized using GeneSpringGX11 software (Agilent Technologies) with default parameters. Differential expression of lncRNAs was screened in genes flagged as “detected” by the software. Differentially expressed genes were identified through the random variance model.
Quantitative polymerase chain reaction
Total mRNA from 31 gastric cancer tissues and 31 corresponding non-tumor tissues was isolated using TRIzol Reagent according to the manufacturer’s protocol. Complementary DNAs from all samples were synthesized from 1.0 μg of total RNA using the PrimeScript® 1st strand cDNA synthesis kit (TaKaRa Bio, Shiga, Japan) following the manufacturer’s protocol. PVT1 expression levels were quantified using a LightCycler Brilliant SYBR Green quantitative reverse transcriptase polymerase chain reaction (PCR) kit (Roche Applied Science, Indianapolis, IN, USA) following the manufacturer’s protocol, with the following primers: forward, TTGGCACATACAGCCATCAT and reverse, GCAGTAAAAGGGGAACACCA. The length of the quantitative PCR product was 102 base pairs. The PVT1 expression level in each sample was normalized to the respective human β-actin expression level, with the following primers: forward, AGCGAGCATCCCCCAAAGTT; and reverse, GGGCACGAAGGCTCATCATT (product length, 285 base pairs). The specificity of each PCR reaction was confirmed by melting curve analysis.
Statistical analysis
The statistical significance of differences between cancerous and adjacent non-cancerous tissues was determined using the Wilcoxon signed-rank test. The correlation between PVT1 expression and patient clinicopathological variables was analyzed by the Fisher’s exact probability test. A P-value <0.05 was regarded as being statistically significant. All statistical analyses were performed using Statistical Package for the Social Sciences version 11.0 software for Windows (SPSS Inc., Chicago, IL, USA).
Results
Microarray analysis
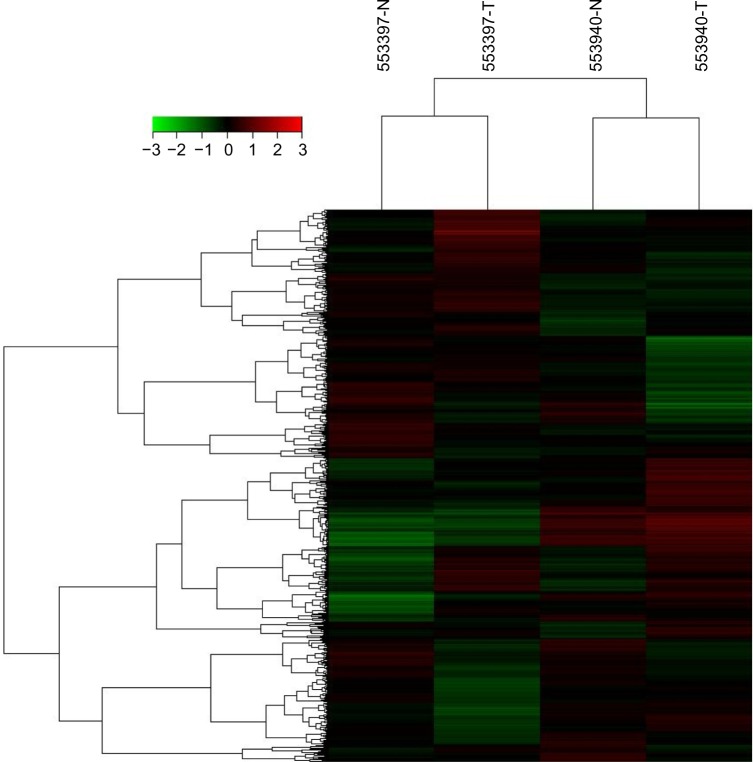
A total of 252 lncRNAs were upregulated in gastric cancer tissues identified from the microarrays. Twenty-eight of the upregulated lncRNAs had an obvious fold change in the gastric cancer tissues when compared with corresponding non-tumor tissues (Figure 1 and Table 1). In total, 545 lncRNAs were upregulated in paclitaxel-resistant SGC7901 cells when compared with normal SGC7901 cells (data supplied). PVT1 expression levels in the gastric cancer tissues were 5.128 and 5.755 times higher than those in the corresponding non-tumor tissues (Table 1). In addition, PVT1 expression was 3.715 times higher in paclitaxel-resistant SGC7901 cells than in normal SGC7901 cells (data supplied). Based on these results and recent discoveries concerning the role of PVT1 in tumorigenesis and resistance to chemotherapy,9,11 we studied the expression of PVT1 further in patients with gastric cancer and its correlation with clinical features.
Figure 1.
Cluster diagram of lncRNA expression data in gastric cancer and corresponding non-tumor tissues detected by the Agilent Human LncRNA 4×180 K Expression Microarray (Agilent Technologies Inc., Santa Clara, CA, USA).
Abbreviation: lncRNA, long-non-coding RNA.
Table 1.
Twenty-eight candidate lncRNAs expressed with fold changes in two gastric cancer tissues, compared with corresponding non-tumor tissues (only the most obvious data are shown for the lncRNAs with different transcripts)
| Fold change of T1 vs N1 | Fold change of T2 vs N2 | Gene biotype | Ensemble gene ID | HGNC symbol |
|---|---|---|---|---|
| 27.20002 | 104.10264 | Antisense | ENSG00000238133 | MLK7-AS1 |
| 16.414074 | 4.819842 | LncRNA | ENSG00000258294 | RP11-314D7.1 |
| 12.021113 | 5.851729 | LncRNA | ENSG00000236039 | AC019117.2 |
| 10.219363 | 5.664894 | LncRNA | ENSG00000241163 | RP11-398A8.3 |
| 8.035696 | 3.0575392 | LncRNA | ENSG00000222041 | LINC00152 |
| 6.8010974 | 2.6033688 | LncRNA | ENSG00000226598 | AC017060.1 |
| 5.912395 | 4.000447 | Antisense | ENSG00000224078 | SNHG14 |
| 5.128479 | 5.754559 | LncRNA | ENSG00000249859 | PVT1 |
| 4.606247 | 4.3609843 | LncRNA | ENSG00000167117 | LINC00483 |
| 3.909583 | 2.458897 | LncRNA | ENSG00000244306 | CTD-2314B22.3 |
| 3.8943083 | 4.077277 | LncRNA | ENSG00000224397 | RP11-290F20.3 |
| 3.698091 | 5.106683 | Antisense | ENSG00000243766 | HOTTIP |
| 3.5476947 | 4.3146453 | Antisense | ENSG00000254560 | RP11-1L12.3 |
| 3.3448012 | 4.3363175 | Antisense | ENSG00000251179 | RP11-893F2.9 |
| 3.2606792 | 2.2400966 | Polymorphic_pseudogene | ENSG00000244682 | FCGR2C |
| 3.0598006 | 3.2264988 | LncRNA | ENSG00000230061 | AP001065.2 |
| 3.0492082 | 3.112142 | LncRNA | ENSG00000233930 | RP13-25N22.1 |
| 2.9770749 | 4.8328776 | LncRNA | ENSG00000227888 | FAM66A |
| 2.8799016 | 2.383093 | Processed transcript | ENSG00000233283 | RP11-357H14.20 |
| 2.8781404 | 2.6564493 | Processed transcript | ENSG00000250610 | RP11-738E22.2 |
| 2.7795174 | 2.031886 | LncRNA | ENSG00000230944 | AC026202.5 |
| 2.7087636 | 25.26169 | LncRNA | ENSG00000204241 | RP11-713P17.3 |
| 2.5824153 | 2.942296 | LncRNA | ENSG00000136315 | RP11-84C10.2 |
| 2.453376 | 2.046279 | LncRNA | ENSG00000214548 | MEG3 |
| 2.4336877 | 3.0326896 | LncRNA | ENSG00000248227 | RP11-83C7.1 |
| 2.2753818 | 2.2845905 | LncRNA | ENSG00000256969 | RP11-320N7.2 |
| 2.1950364 | 4.0923414 | LncRNA | ENSG00000245975 | RP11-30K9.6 |
| 2.125026 | 2.8170245 | LncRNA | ENSG00000235572 | RP11-555H7.2 |
Abbreviations: HGNC, Human Genome Nomenclature Committee; lncRNAs, long-noncoding RNAs.
PVT1 expression
PVT1 expression levels in cancerous and adjacent noncancerous tissues from patients with gastric cancer were examined by quantitative reverse transcriptase PCR and normalized by β-actin expression levels. The Wilcoxon signed-rank test showed that PVT1 expression was significantly higher in cancerous tissues than in adjacent noncancerous tissues (P=0.0414).
PVT1 expression and clinical features
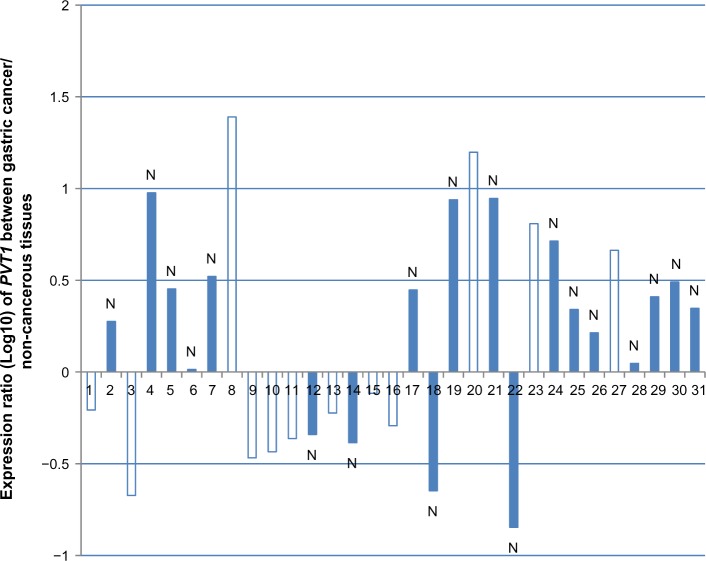
In Figure 2 and Table 2, the human gastric cancer tissues are classified according to PVT1 expression level into a high PVT1 expression group (PVT1 in cancer tissue/PVT1 in non-cancerous tissue >1.0) and a low PVT1 expression group (PVT1 in cancer tissue/PVT1 in non-cancerous tissue <1.0). The association of PVT1 expression with the clinicopathological features of patients with intestinal-type gastric cancer is summarized in Table 2. A significant association was found between PVT1 expression and lymph node metastasis (P=0.022). No correlation was found between PVT1 expression and patient age, sex, tumor metastasis, or stage of gastric cancer.
Figure 2.
Ratio of relative PVT1 expression in cancerous tissues versus adjacent noncancerous tissues from 31 patients with gastric cancer, examined by quantitative reverse transcriptase polymerase chain reaction. N indicates the number of patients with lymph node metastasis.
Table 2.
Association between PVT1 expression and clinicopathological factors in human gastric cancers
| High PVT1 expression | Low PVT1 expression | P-valuea | |
|---|---|---|---|
| Age | 60.7895±7.0441 | 64.7500±9.0566 | 0.1827 |
| Sex | 0.1285 | ||
| Male | 11 | 10 | |
| Female | 8 | 2 | |
| T | 1.0000 | ||
| T1, T2 | 4 | 3 | |
| T3, T4 | 15 | 9 | |
| N | 0.0220 | ||
| N0 | 4 | 8 | |
| N1, N2, N3 | 15 | 4 | |
| M | 0.2645 | ||
| M0 | 16 | 12 | |
| M1 | 3 | 0 | |
| Stage | 0.0596 | ||
| 1, 2 | 5 | 8 | |
| 3, 4 | 14 | 4 |
Note:
Fisher’s exact probability test.
Discussion
A set of lncRNAs showed differential expression between human gastric cancer and non-cancerous tissue. Thus, these lncRNAs may play important roles in the carcinogenesis and invasiveness of human gastric cancer and are potential candidates for new biomarkers and therapeutic targets.6–9
PVT1, one such oncogenic lncRNA, is a large (>300 kb) locus located 57 kb downstream of MYC on human chromosome 8q24.12–15 Although no protein product or regulatory RNA has yet been identified among PVT1 transcripts, the importance of this locus has caught the attention of many researchers, as it is a site of both tumorigenic translocations and retroviral insertions. Translocation breakpoints within either the MYC or PVT1 locus are responsible for the characteristic lesions associated with Burkitt lymphoma and mouse plasmacytomas.13–18
Coamplification of human MYC and PVT1 has been shown to correlate with rapid progression of breast cancer as well as poor clinical survival in postmenopausal women and in patients with HER2-positive breast cancer.19 Recent studies have discovered a cluster of miRNAs within the PVT1 genomic DNA region, including miR1204, miR1205, miR1206, miR1207, and miR1208.20,21 The regulatory functions of these miRNAs within MYC-PVT1 locus are complicated, only the function of miR1204 has been partially proved. MiR1204 plays an antiproliferative role, inducing cell death or cell cycle arrest, as a functional target of p53 at the PVT1 locus. MiR1204 expression can also increase p53 levels, suggesting there is a positive feedback between miR-1204 and p53.22 So far, at least one PVT1 transcript microRNA product, miR1204, has been shown to be overexpressed and is thus defined as oncogenic.9,22 These additional non-coding RNA transcripts, exonic PVT1 sequences, and other miRs are now thought to be either transcriptional byproducts of miR1204 regulation or additional functional targets, but no further evidence is provided. So, except for two aspects of the mechanism of PVT1 in the prognosis of cancer (including a positive feedback regulation with p53 and coamplification with MYC), more studies need to be directed to the complex networks between PVT1, p53, MYC, and other genes.
In this study, we examined the expression of lncRNAs with 77×103 probes from the Agilent Human 4×180 K Microarray to identify specific lncRNA markers for human gastric cancers at the gene level. These probes correspond to 37,000 lncRNAs and 34,000 mRNAs. We also compared lncRNA expression between an SGC7901 human gastric cancer cell line and a paclitaxel-resistant SGC7901 cell line. The differential expression observed may reveal the mechanism of and biomarkers for paclitaxel resistance in human gastric cancer. Based on the microarray results and in light of recent reports,11,13–19 we ranked PVT1 as the lncRNA with the highest potential as a biomarker and a therapeutic target for gastric cancer.
In order to compensate for the deficiency of clinical cases in the microarray analysis and to develop a convenient and economical method for large-scale clinical studies, we used the quantitative PCR technique to confirm the relevance of PVT1 as a biomarker for gastric cancer. Our results clearly show that PVT1 expression is significantly enhanced in gastric cancer tissues when compared with non-cancerous tissues of the stomach. A high level of PVT1 expression was also associated with lymph node metastasis in patients with gastric cancer. No correlation was found between PVT1 expression and age, sex, tumor, metastasis, or stage of gastric cancer.
In conclusion, we demonstrated that PVT1 is highly expressed in human gastric cancer. Enhanced expression of PVT1 was associated with invasion to the lymph nodes and with resistance to paclitaxel. Therefore, these results indicate that PVT1 is a potential candidate as a biomarker for gastric cancer. Screening for PVT1 expression could potentially improve early diagnosis, therapy, and drug resistance, and thus improve quality of life for patients with gastric cancer.
Acknowledgments
The study was supported by the China National Science Foundation (81300321), the Fujian Provincial Natural Science Fund (2013J01369), and the Fujian Provincial Natural Science Fund (2014J01419).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(Suppl 8):S4–S66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM. International variation. Oncogene. 2004;23(38):6329–6340. doi: 10.1038/sj.onc.1207726. [DOI] [PubMed] [Google Scholar]
- 3.Stewart BW, Kleihues P. World Cancer Report. Lyon, France: IARC Press; 2003. [Google Scholar]
- 4.Perkel JM. Visiting “noncodarnia”. Biotechniques. 2013;54(6):301, 303–304. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- 5.ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Yang F, Yuan JH, et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34(3):577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 8.Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 9.Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. doi: 10.1186/1742-4690-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton MA, Russo RC, Thurston RV. Trimmed Spearman-Kärber CPE method for estimating median lethal concentrations in toxicity bioassays. Environ Sci Technol. 1977;13:714–719. [Google Scholar]
- 11.You L, Chang D, Du HZ, Zhao YP. Genome-wide screen identifies PVT1 as a regulator of gemcitabine sensitivity in human pancreatic cancer cells. Biochem Biophys Res Commun. 2011;407(1):1–6. doi: 10.1016/j.bbrc.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 12.Shtivelman E, Henglein B, Groitl P, Lipp M, Bishop JM. Identification of a human transcription unit affected by the variant chromosomal translocations 2;8 and 8;22 of Burkitt lymphoma. Proc Natl Acad Sci U S A. 1989;86(9):3257–3260. doi: 10.1073/pnas.86.9.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huppi K, Siwarski D, Skurla R, et al. Pvt-1 transcripts are found in normal tissues and are altered by reciprocal (6;15) translocations in mouse plasmacytomas. Proc Natl Acad Sci U S A. 1990;87(18):6964–6968. doi: 10.1073/pnas.87.18.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shtivelman E, Bishop JM. Effects of translocations on transcription from PVT. Mol Cell Biol. 1990;10(4):1835–1839. doi: 10.1128/mcb.10.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shtivelman E, Bishop JM. The PVT gene frequently amplifies with MYC in tumor cells. Mol Cell Biol. 1989;9(3):1148–1154. doi: 10.1128/mcb.9.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pleasance ED, Stephens PJ, O’Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463(7278):184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pomerantz MM, Beckwith CA, Regan MM, et al. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 2009;69(13):5568–5574. doi: 10.1158/0008-5472.CAN-09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enciso-Mora V, Broderick P, Ma Y, et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3) Nat Genet. 2010;42(12):1126–1130. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borg A, Baldetorp B, Fernö M, Olsson H, Sigurdsson H. c-Myc amplification is an independent prognostic factor in postmenopausal breast cancer. Int J Cancer. 1992;51(5):687–691. doi: 10.1002/ijc.2910510504. [DOI] [PubMed] [Google Scholar]
- 20.Huppi K, Volfovsky N, Runfola T, et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. 2008;6(2):212–221. doi: 10.1158/1541-7786.MCR-07-0105. [DOI] [PubMed] [Google Scholar]
- 21.Bisio A, De Sanctis V, Del Vescovo V, et al. Identification of new p53 target microRNAs by bioinformatics and functional analysis. BMC Cancer. 2013;13:552. doi: 10.1186/1471-2407-13-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barsotti AM, Beckerman R, Laptenko O, Huppi K, Caplen NJ, Prives C. p53-dependent induction of PVT1 and miR-1204. J Biol Chem. 2012;287(4):2509–2519. doi: 10.1074/jbc.M111.322875. [DOI] [PMC free article] [PubMed] [Google Scholar]