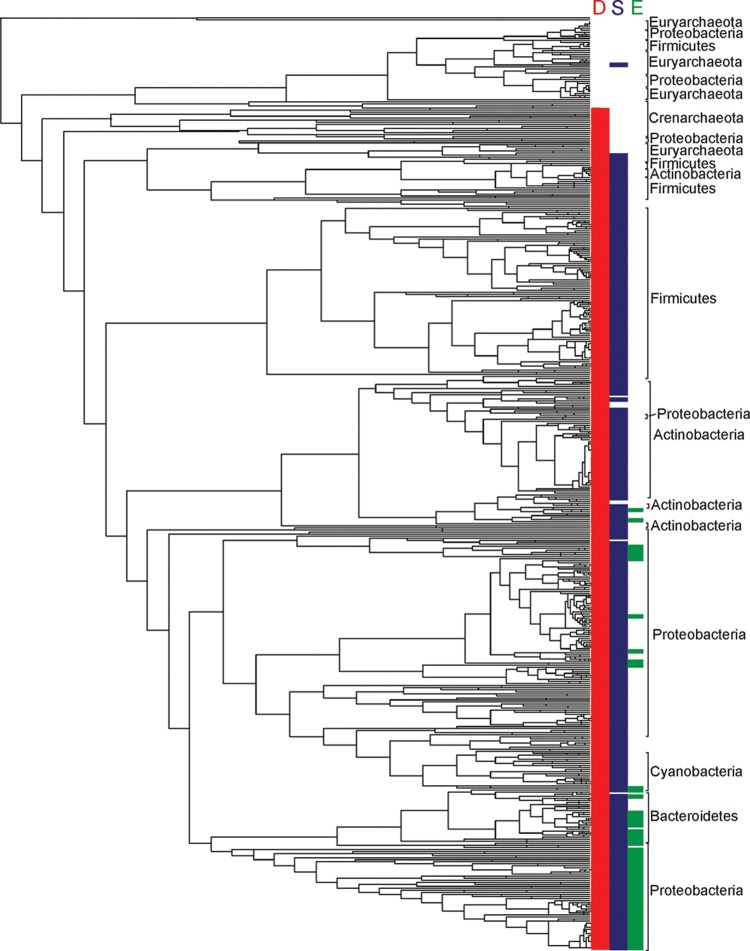
Figure 5.
Rate-smoothed phylogenetic reconstruction of concatenated SufBC alignment blocks from 1094 bacterial and archaeal taxa (eukarya excluded). Representative SufB and SufC were identified in available genome sequences in October 2011 and were aligned individually with ClustalW103 specifying default alignment parameters. Paralogs of SufB (i.e., SufD) and SufC (i.e., other members of the ABC transporter ATP-binding protein family) were included in each alignment, respectively, and were later used to root the phylogeny. SufB and SufC alignment blocks were concatenated with PAUP (version 4.0, Sinauer Associates, Inc.). The maximum likelihood phylogenetic reconstruction was inferred using PhyML version 3.0 specifying the LG substitution matrix and a four-category gamma substitution model.104 The concatenated SufBC phylogeny was rate-smoothed using the penalized-likelihood approach105 as implemented by the “chronopl” command in the R statistical package (version 2.15.0) Picante (version 1.3)106 specifying a smoothing parameter of 1.0. The distribution of SufD (red), SufS (green), and SufE (blue) in the suf operon in each taxon was mapped on the phylogeny using base commands in the Picante package within R. The primary taxonomic rank at the phylum level is indicated for major clusters.