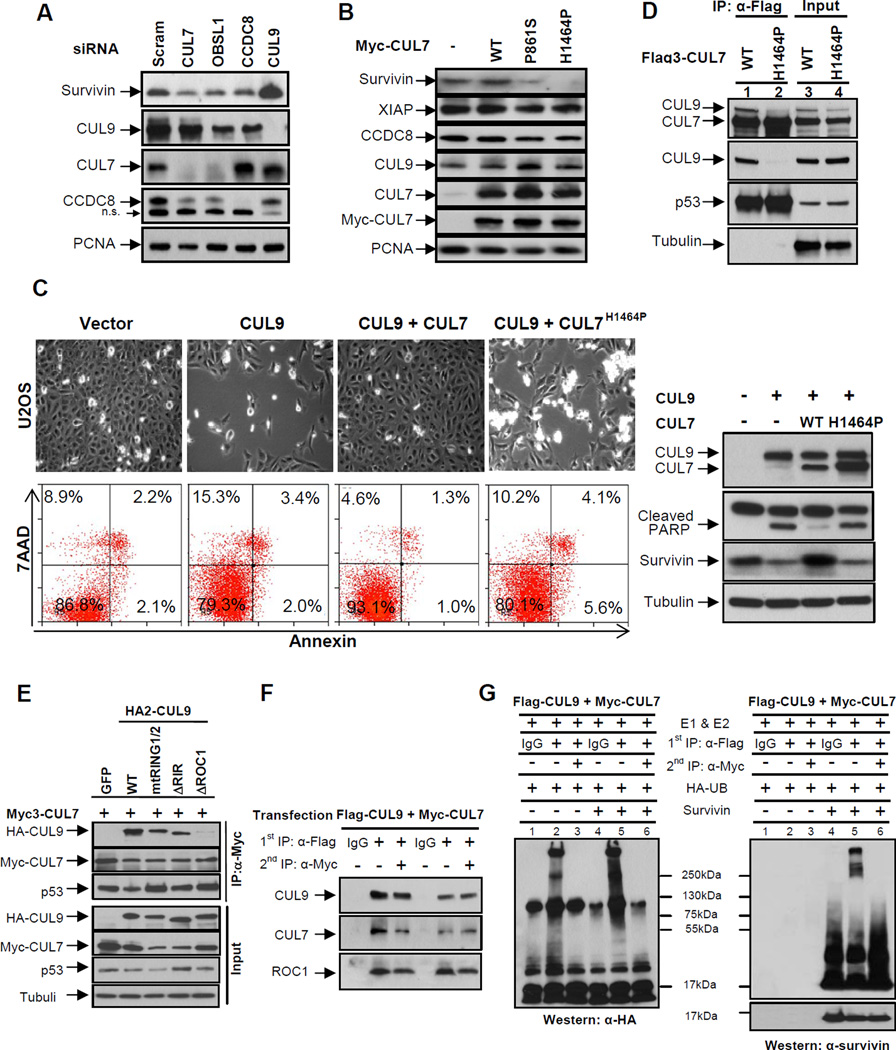
Figure 6. CUL7 binds to and inhibits CUL9.
(A) U2OS cells were transfected with indicated siRNA oligos, and 48 hours following transfection, the steady state levels of indicated proteins were determined by immunoblotting.
(B) U2OS cells were transduced with retroviruses expressing indicated wild-type or mutant CUL7 proteins. The steady state levels of proteins were determined by immunoblotting.
(C) U2OS cells were transfected with indicated plasmids for 48 hours, a representative phase contrast view of each sample is shown. The cells were then collected, stained with Annexin V and 7AAD, and then analyzed by FACS. Protein expression was determined by immunoblotting.
(D) Lysates prepared from U2OS cells stably expressing wild-type or mutant CUL7 were immunoprecipitated with anti-Flag antibody, followed by immunoblotting with indicated antibodies to assess CUL7-CUL9 interaction; α-tubulin served as a loading control.
(E) Lysates from transfected U2OS cells were immunoprecipitated with anti-Myc3 antibody, followed by immunoblotting with antibodies against CUL9, CUL7 and p53. See also Figure S5.
(F) Lysates from U2OS cells co-transfected with Flag3-CUL9 and Myc-CUL7 were subjected to sequential IP to assess the direct binding requirement for CUL9 inhibition by CUL7. Flag and Myc antibodies were used for the first IP (lanes 2 and 5) and second IP (lanes 3 and 6), respectively.
(G) Sequential immunoprecipitates from (F) were used for assaying survivin ubiquitylation in vitro. Reactions were resolved by SDS-PAGE and probed with either antibody to HA-ubiquitin or survivin.