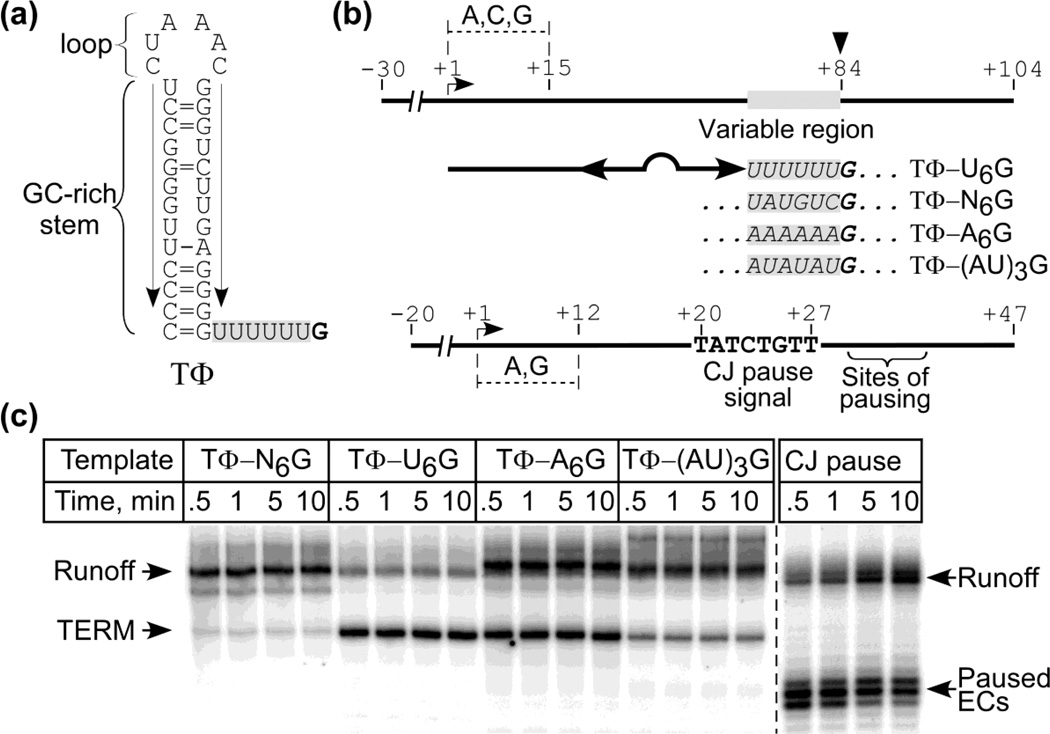
Figure 3. Termination at stem-loop poly(A) signals.
Panel (a). Predicted organization of TΦ. Arrows indicate the ascending and descending arms of the stem; the U6 tract downstream is shaded, and the site of termination (G) is in bold.
Panel (b). Template design. Templates encode transcripts having the sequence found in the stem-loop structure in TΦ (double headed arrow) followed by a variable region (boxed sequence); the site of termination is at +84. The signals are identified at the right by noting the nature of the variable region. The lower scheme explains the design of a template containing a CJ pause signal.
Panel (c). Termination at TΦ and its variants. Start up complexes halted at +15 in the presence of GTP, ATP and CTP for 5 min were mixed with a 20-fold excess of promoter trap (to bind free RNAP and prevent reinitiation) and α-[32P]-UTP (to allow extension of the transcripts and subsequent detection). The reactions were stopped after the time intervals indicated and the products were resolved by electrophoresis; positions of terminated and runoff products are indicated to the left. A similar experiment was carried out with a template that contains a sequence-specific pause signal for T7 RNAP found in the concatamer junction (CJ) of replicating T7 DNA29 (right side of panel).