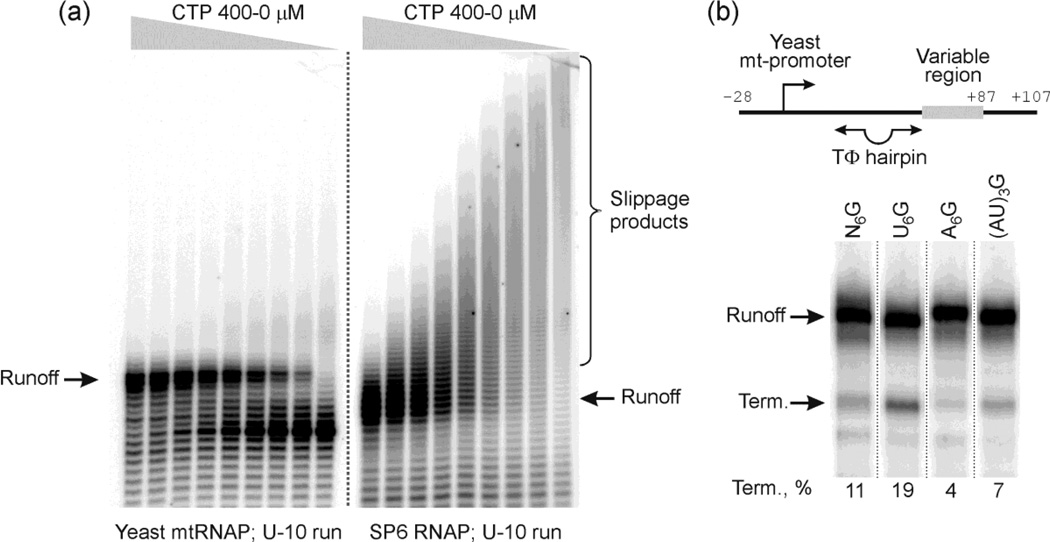
Figure 4. The single-subunit yeast mitochondrial RNAP resists slippage on homopolymeric tracts.
Panel (a). Slippage by mtRNAP and SP6 RNAP. Transcription by mtRNAP (left side) and SP6 RNAP (right side) was carried out for 20 min at 37 °C in the presence of ATP, GTP, UTP, α-[32P]-GTP, and CTP added to concentrations of 400, 200, 100, 50, 25, 13, 6, 3, and 0 µM (left to right). The templates encoding U10 runs in the variable region were prepared as in Fig. 2a except a consensus yeast mitochondrial promoter or SP6 promoter were provided for corresponding enzymes. The transcripts formed were resolved by 16 % denaturing PAGE; expected run-off and slippage products are indicated.
Panel (b). Termination by mtRNAP. Templates carrying a WT and altered TΦ termination signal were prepared as in Fig. 3b except a consensus mitochondrial promoter was used upstream of the signals instead of a T7 promoter (see scheme on top). Multiple round transcription reactions with the templates indicated were performed in the presence of Rpo41p (mtRNAP), Mtf1p, ATP, CTP, GTP, UTP, and α-[32P]-GTP in transcription buffer for 15 min at 30 °C and resolved by 20 % PAGE. Run-off transcripts and termination products are marked on the left and the termination efficiency is indicated under each lane.