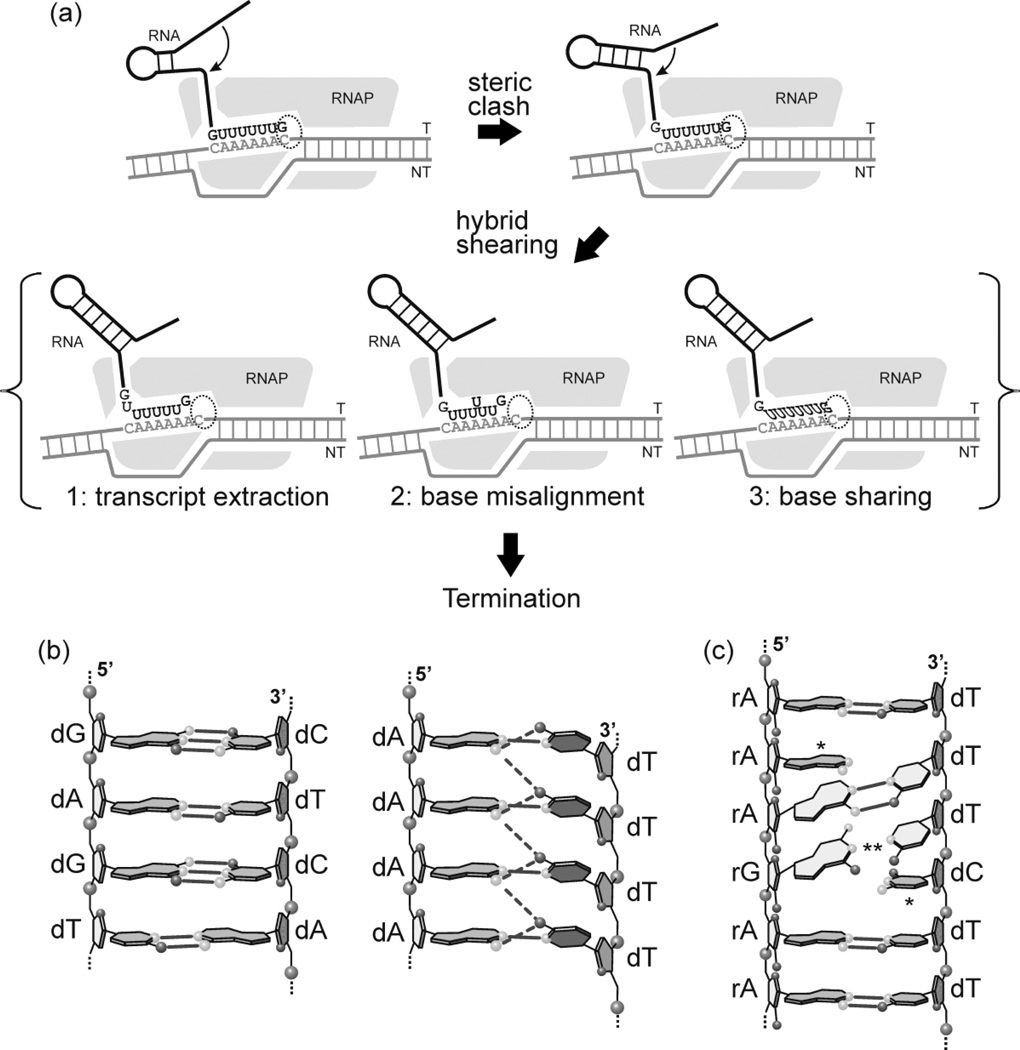
Figure 6. A model for termination by T7 RNAP.
Panel (a). Model for the early stages of the termination process. As the RNAP (gray shape) transcribes through the termination signal, annealing of the stem-loop region of the transcript results in steric clash with the exit pore, generating a shearing force that results in a change in the organization of the active site (dashed oval) and the formation of a “locked” complex that represents an early step in the termination pathway. The transmission of the shearing force along the hybrid is facilitated by the presence of homopolymeric tracts that promote transcript slippage under passive conditions. While active slippage (i.e., additional incorporation of UMP residues) is not involved in this model (and in fact, would be expected to decrease the efficiency of termination due to a relief in steric clash, see Discussion) the presence of slippage-prone sequences lowers the thermodynamic barrier to changes in the hybrid. We do not know the organization of the hybrid in such a complex nor the mechanism by which the shearing force is transmitted. A number of pathways have been proposed (for review see1). In scheme 1, extraction of the transcript removes the 3’ end from the active site; the formation of a mismatched base pair may limit extraction to one or a few base pairs. In scheme 2, base misalignment along the hybrid results in extraction of the 3’ end from the active site. In scheme 3, (base sharing, see below) changes in the geometry of the hybrid, without slippage, alter the orientation of the 3’ end of the transcript in the active site.
Panel (b). Base sharing. Cross strand hydrogen bonds may facilitate transcript slippage by base sharing (see1 for review). Structural data indicate that A:T base pairs in homopolymeric (dA:dT)n tracts exhibit a high propeller twist that may be stabilized by cross strand hydrogen bonds between the adenine N-6 amine of a base pair on one strand and the thymine O–4 of the succeeding base pair on the opposite strand (right side)49 This is not observed in non-homopolymeric tracts (left side).
Panel (c). Alternate RNA:DNA hybrid structures; unzipping and slippage of the RNA:DNA hybrid in the polypurine tract of an HIV-RT complex. Structural analysis of HIV-RT complexes in association with the “polypurine tract” reveals a non-canonical form of the RNA:DNA hybrid that involves both slipped and mismatchedbases44. Misaligned (*) and mismatched (**) bases are indicated.