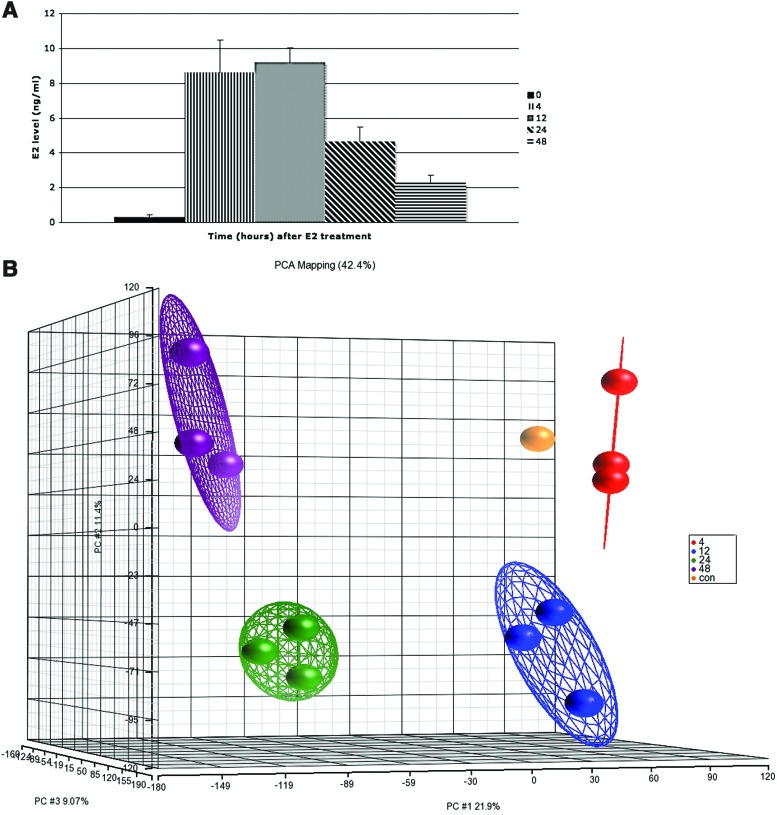
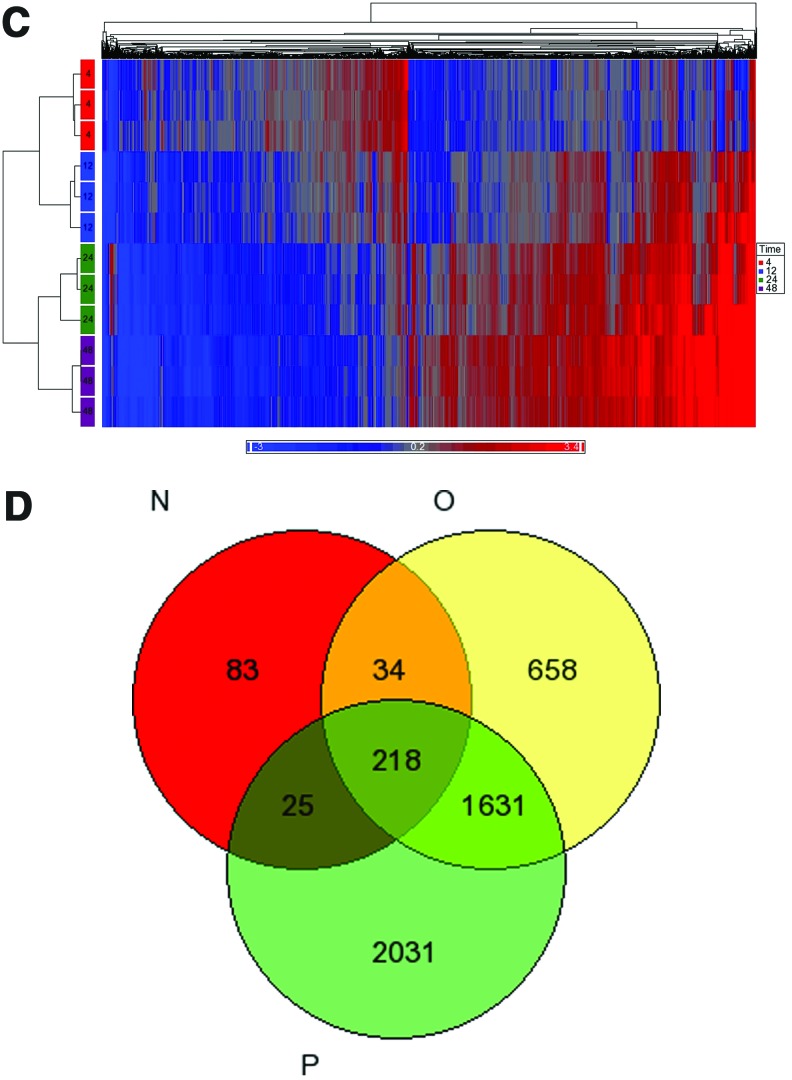
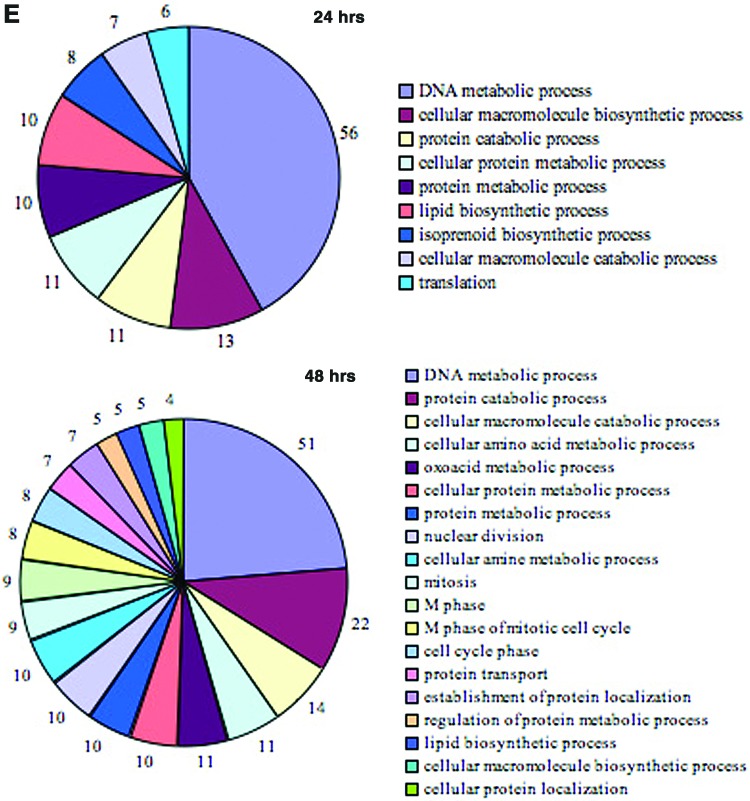
FIG. 1.
Alterations in gene expression profiles in the zebrafish liver after E2 treatment. (A) E2 concentrations in the plasma of control and E2-treated fish (mean±standard error of the mean [SEM], n=3). (B) Principal component analysis (PCA) plot of gene expression microarray results after E2 treatment. Each dot represents a chip of the different time points (4, 12, 24, 48 hrs). Another chip hybridized two control pools together and served as a control chip in the PCA plot. Performed using Partek-Genomics-Suite software. (C) Heatmap representation of regulated genes at different time points after E2 treatment. The range of expression values is from −3-fold to +3-fold. The rectangles on the left represent chips. Microarray results were hierarchically clustered using Partek-Genomics-Suite software. (D) Venn diagram of differentially expressed genes of gene expression microarrays after E2 treatment: 12 h (N), 24 h (O), 48 h (P). (E) Enriched gene ontology terms in gene expression microarray results after E2 treatment, as were identified using the DAVID functional annotation tool. Shown are significant-log2 Benjamini p-values of biological process terms (level 4) of differentially expressed genes at 24 and 48 h. Color images available online at www.liebertpub.com/zeb