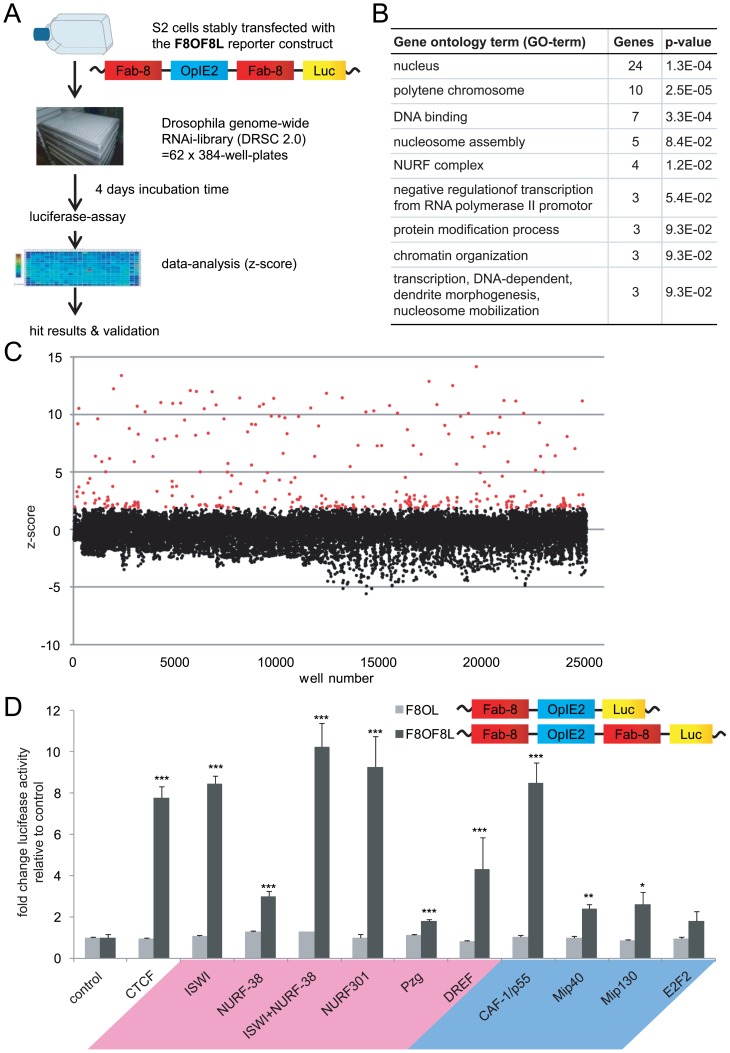
Figure 1. RNA interference (RNAi) identifies 78 factors inducing insulator reporter gene activity including NURF and dREAM components.
(A) Workflow of the RNAi screen in 66×384-well plates from the DRSC. Knockdown of 13900 genes was done with Drosophila S2 cells with the integrated F8OF8L insulator reporter construct (F8, Fab-8; O, OpIE2 enhancer; L, luciferase). (B) Top GO-terms (determined via GeneCodis [58]–[60]) for the 78 identified genes. (C) High-throughput data shown in a dotplot diagram. Z-scores are indicated for every well (well number). For many gene products several wells contain different dsRNA sequences targeting the same gene. Z-scores higher than two are highlighted in red. (D) Individual depletion of NURF and dREAM components and associated factors verify enhancer blocking function. S2 cell pools with the integrated F8OF8L insulator reporter (dark grey) or the control F8OL reporter construct (light grey) were incubated with dsRNA against factors of the NURF-complex (pink): ISWI, NURF-38, CAF1/p55, NURF301, Pzg, DREF or against the dREAM-complex (blue): CAF1/p55, Mip40, Mip130, E2F2. Reporter gene activity is expressed as fold change relative to control knockdown. Error bars indicate the standard deviation of three individual replicates. (p-values: *≤0.05, **≤0.01, ***≤0.001).