Abstract
AIM: To evaluate the receptor protein which can specifically bind to β2GP I on the membrane of hepatocellular carcinoma (HCC) cell line SMMC-7721, and to study the biological function of the receptor.
METHODS: Through β2GP I -affinity chromatography column, the peptid-polysome-mRNA complex, which can specially bind to β2GP I , stayed with the column and was separated from the whole polysome of liver cells, and then eluted and collected. Using cDNA synthesis kit and cDNA PCR kit, the corresponding cDNA was obtained and sequenced. RT-PCR was used to amplify annexin II , and flow cytometry was used to study the competitive binding of annexin II with β2GP I to SMMC-7721.
RESULTS: A total of 1.1 kb of the cDNA fragment of the specific binding protein of β2GP I on liver cell membrane was obtained. The sequence of cDNA shared high homology with human annexin II (98%). Annexin II was expressed on the membrane of SMMC-7721, and could compete with β2GP I for combining with SMMC-7721.
CONCLUSION: The receptor for β2GP I on membrane of SMMC-7721 cells is annexin II , which might bridge HBV to infect hepatocytes.
Keywords: β2-GlycoproteinI; Hepatocellular carcinoma cell, Human annexin II
INTRODUCTION
β2-GlycoproteinI(β2GP I ) is an abundant plasma glycoprotein. Because of its high affinity binding with plasma phosphorlipid, it is also named apolipoprotein H. It has been shown that β2GP I could act as anticoagulant[1] and is an important autoantigen in the antiphospholipid antibody syndrome[2,3]. Up to now, it has been found that β2GP I has many other functions. In 1994, Mehdi et at[4] demonstrated that β2GP I was capable of binding to recombinant hepatitis B surface antigen (rHBsAg), suggesting that β2GP I may facilitate entry of the virus into hepatocytes. They also found that rHBsAg bound to β2GP I very poorly if β2GP I was coated directly on a microtiter well, or if it was presented in a soluble form. While binding was 100-fold more efficient when β2GP I was presented as a complex with monoclonal antibody (mAb) P2D4. These results suggest that chemical modification of β2GP I makes it highly reactive with rHBsAg[5]. Recently it has been reported that β2GP I -HBsAg combining was relative to the presence of hepatitis B virus markers and β2GP I binding activity for HBsAg was higher in sera from patients in the active virus replication phase[6].
We have previously finished purification and evaluation of formation of β2GP I , and found that the level of anti-β2GP I antibodies in patients with chronic hepatitis B and post-hepatitis B cirrhosis was significantly increased, thereby suggesting that β2GP I can take part in HBV infection. In our previous studies, we also verified that β2GP I could specifically bind to rHBsAg[7,8]. Through ligand blot analysis, fluorescence microscope and flow cytometry, it was probably the first time to prove that there exists a protein on SMMC-7721 cell membrane that can bind to β2GP I with specificity[9]. We concluded that the protein may be the receptor of β2GP I , and it might be a carrier which can bridge HBV to invade hepatocytes. In this study, we will evaluate the receptor on SMMC-7721 cell line that can bind to β2GP I with specificity.
MATERIALS AND METHODS
Preparation of β2GP I -affinity chromatography column
Purified β2GP I was preserved in our laboratory. According to instructions of Epoxy-activated Sepharose 6B, the medium was suspended in distilled water. The purified β2GP I was dissolved in buffer solution, mixed with gel granules for 16 h with shake cultivation at 37°C, and then was packed in column. The column was washed with buffer solution, distilled water, buffer solution A and buffer solution B by turns in order to eliminate excess β2GP I . The remained active radical was blocked with 1 mol/L ethanolamine at 37°C. Protein in the collected elutriant was quantitated with BCA methods.
Extraction of polysome of liver cells
All procedures were performed at 4°C. Under aseptic conditions, connective tissue and fat was eliminated from liver with scissors. Then the liver tissue was cut into scraps and grinded in cell homogenizer. After being filtered through stainless steel screen, the products were washed with solution A (pH 8.0, 60 mmol/L sodium phosphate buffer solution, 45 mmol/L NaCl, 55 mmol/L glycose, 1 μg/mL cycloheximide). The liver cells were counted, and 2 × 1010 cells were resuspended in solution B (pH 8.0, 60 mmol/L sodium phosphate buffer solution, 45 mmol/L NaCl, 55 mmol/L glycose, 1 μg/mL cycloheximide, 40 U/mL heparin, 10 mmol/L DTT). Then 10 mL of solution C (pH 7.8, 50 mmol/L Tris-HCl, 300 mmol/L NaCl, 10 mmol/L MgCl2, 1 μg/mL cycloheximide, 20 U/mL heparin) and nonidet P-40 were added slowly to a final concentration of 3.5 mL/L in order to split them. After incubation for 5 min and centrifugation, the supernatant was collected. Sodium deoxycholate of final concentration of 3.5 mL/L was added to destroy microsome completely. Remainder of supernatant was loaded at 65% (W/W) sucrose in Buffer D (pH 7.6, 25 mmol/L Tris-HCl, 150 mmol/L NaCl, 5 mmol/L MgCl2, 20 U/mL heparin), then ultracentrifuged at 5000 r/min for 2 h. The deposition was resuspended in solution E (pH 7.6, 25 mmol/L Tris-HCl, 150 mmol/L NaCl, 5 mmol/L MgCl2, 1 mL/L nonidet P-40, 1 μg/mL cycloheximide, 20 U/mL heparin), and stored in liquid N2.
Affinity purification of mRNA of the receptor protein
According to manufacturer’s instructions, the β2GP I -affinity chromatography column was balanced with solution A. The polysome extraction was slowly loaded on the column repeatedly. After being processed for 1 h, the column was washed with 20-30 volume solution E, eluted with solution F (pH 7.6, 25 mmol/L Tris-HCl, 150 mmol/L EDTA, 20 U/mL heparin) in order to collect the objective mRNA which can specifically bind β2GP I , and then 0.5 mol/L NaCl and 1 g/L SDS were added. The above solution was loaded on oligo-dT cellulose column. The oligo-dT cellulose column was washed with solution G (pH 7.6, 25 mmol/L Tris-HCl, 500 mmol/L NaCl, 1 g/L SDS, 20 U/mL heparin), then eluted with solution G without NaCl. The elutriant containing objective mRNA was concentrated, washed, dissolved in sterile water, and stored at -80°C.
Synthesis of double strands cDNA
Synthesis of 1st strand cDNA: mRNA was treated at 65°C for 5 min, and ice bathed immediately. A total volume of 10 μL of reaction mixture contained 2 μL of sample mRNA, 2 μL of 5 × 1st Strand Synthesis buffer, 1 μL of dNTP, 1 μL of RNase inhibitor, 1 μL of oligo dT-RA primer, 1 μL of RAV-2 reverse transcriptase, 2 μL of DEPC H2O. The reverse transcription was performed for 10 min at 30°C, for 1 h at 42°C, and finally for 5 min at 80°C.
Synthesis and external smoothing of 2nd strand cDNA: In aforementioned reaction mixture, 10 μL of 5 × 2nd Strand Synthesis buffer, 20.5 μL of DEPC H2O and 1.0 μL of E. coli DNA Ligase Mixture were added and mixed gently. After incubation for 1 h at 12°C, 1 h at 22°C, and 10 min at 70°C in turn, the mixture was mixed gently with 2 μL of T4 DNA PolyMeraseIand kept at 37°C for 10 min. Then 4 μL of Stop Solution was added to stop the reaction.
Purification of double strand cDNA: The obtained cDNA was extracted with phenylic alcohol/chloroform, precipitated with isopropanol, washed with ethanol and dried.
Ligation of cassete adaptor
After being dissolved in 5 μL of sterilized water, the above sediment was mixed with 2 μL CA cassette adaptor and 6 μL of ligation solution gently. The reaction remained at 16°C for 30 min. Then 25 μL of 4 mol/L ammonium acetate was added. The products was precipitated with isopropanol and then dissolved in 30 μL of sterilized water at -80°C.
PCR of the double strand cDNA
A total volume of 50 μL of PCR mixture contained 30 μL of prepared cDNA solution, 5 μL of 10 × Ex Taq buffer, 4 μL of dNTP mixture, 0.5 μL of RA primer, 0.5 μL of CA primer, 0.25 μL of Takara Ex Taq and 9.75 μL of sterilized water. PCR mixture was subjected to pre-denaturation at 94°C for 1 min, followed by 35 amplification cycles of denaturation at 94°C for 30 s, primer annealing at 60°C for 30 s and extension at 72°C for 3 min. Finally, the PCR mixture incubated at 72°C for 5 min, reaction was held at 4°C.
Sequencing
According to molecular clone, the PCR products were purified and then linked to TA vector. The recombined plasmid was transfected into receptive E. coli DH5α. Through blue-white screening, the positive colony was obtained. Using plasmid extraction kit, the recombined plasmid was extracted, and sequenced in the company. Finally, the sequence was analyzed using BLAST.
Amplification of annexin II from liver tissue by RT-PCR
According to manufacturer’s instruction (Invitrogen Company), using Trizol reagent, total RNA was isolated from cultured 1 × 107 SMMC-7721 cells and stored at -70°C. In accordance with the instructions of AMV reverse transcriptase, 20 μL of mixture, containing 20 μg of total RNA, 2 μL of oligo (dT) and 18 μL of DEPC H2O, was kept in a 200-μL micro-centrifuge tube, mixed gently and then heated at 70°C for 5 min. After immediate cooling on ice for at least 5 min, 5 μL of dNTP mixture, 10 μL of AMV 5 × buffer, 2 μL of Rnasin, 3 μL of AMV reverse transcriptase and 10 μL of DEPC H2O were added to reach a total volume of 50 μL, followed by incubation at 42°C for 90 min, and then for 5 min at 95°C for inactivation. Thus the RNA was reverse transcribed into cDNA which was stored at -70°C. Referring to mRNA of annexin II from GenBank and following principle of design for primers, a pair of primers was designed: AAAAGATCTCCAGCTTCCTTCAAA (sense); AAAGTCGACATTTCTGGACGCTCA (anti-sense). The reaction system, containing 5 μL of 10 × buffer, 5 μL of Mg2+, 4 μL of dNTP, 1 μL of each sense and antisense primers, 10 μL of cDNA, 1 μL of Taq enzyme and 23 μL of DEPC H2O, was subjected for 40 amplification cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 2 min, followed by a final extension at 72°C for 7 min. Ten microliters of PCR product was electrophoresed on 10 g/L agarose gel.
Flow cytometry
Green fluorescent protein (GFP) and β2GP I labeled with GFP (GFP-β2GP I ) were gifted by Central Laboratory of our hospital. SMMC-7721 cells are preserved in our laboratory. About 9 × 106 of SMMC-7721 cells were collected, washed and divided averagely into 9 tubes. These 9 tubes were randomly divided into three groups, 3 tubes in each group: group A, group B and group C. After being washed twice with PBS, each tube of cells was added with 2 mL of BD FACS Permeabilizing Solution and kept at room temperature for 10 min. Then they were centrifuged to discard the supernatant. The cells were washed with PBS again, and added with 0.45 mL of buffer solution containing Ca2+. Thereafter, groups A, B and C were added with 0 μL, 0 μL and 10 μL of annexin II , respectively and kept at room temperature for 30 min. Finally, 0.05 mL of GFP was added to the each tube of group A, while 0.05 mL of GFP-β2GP I to each tube of groups B and C. All tubes were kept at room temperature for 2 h and washed twice with PBS, and then cells of each tube were suspended in 0.4 mL of PBS and detected using flow cytometry.
RESULTS
Couple rate of prepared β2GP I -affinity chromatography column
Ten milligrams of purified β2GP I was dissolved in couple solution and mixed with gel granule for 16 h with shake cultivation at 37°C. The excess β2GP I was washed with couple solution, distilled water, buffer solution A and buffer solution B by turns and then was collected. The quantity of excess protein in the eluted solution was measured, which was 1.1 mg. So the couple rate was 89%. The couple efficiency was good enough to be used for the latter study.
Electrophoresis of RT-PCR products
With the β2GP I -affinity chromatography column prepared by ourselves, the peptid-polysome-mRNA which can specially bind to β2GP I conjugated with β2GP I on the column, and then was eluted. Through cDNA synthesis kit, the first strand cDNA was obtained. Then the double strand cDNA was acquired. Agar gel electrophoresis confirmed that the molecular weight of the cDNA fragment was 1.1 kb (Figure 1).
Figure 1.
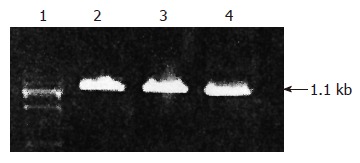
Agarose gel electrophoresis of RT-PCR products. Lane 1: DNA maker; Lanes 2, 3, 4: PCR products.
Gene sequencing and analysis of β2GP I -binding receptor
The recombined plasmid purified from positive cloned bacteria was sequenced in Beijing Dingguo Biological Technique Company. Then the sequence was analyzed for its homology with GenBank BLAST. The results showed that the sequence of cDNA shared high homology with human annexin II (98%) (Figure 2).
Figure 2.

Comparison of the cDNA sequence of receptor of β2GP I with annexin II .
RT-PCR of annexin II
RT-PCR was used to detect the expressions of β-actin and annexin II in HUVEC, SMMC-7721 and hepatoma tissue (Figure 3). Lanes 3, 5 and 7 show β-actin expression. It showed that the total mRNA of the three groups had been extracted successfully. Lanes 2, 4 and 6 show a single band of 1000 bp which represents annexin II . Similarly, HUVEC, SMMC-7721 and hepatoma tissue expressed annexin II .
Figure 3.
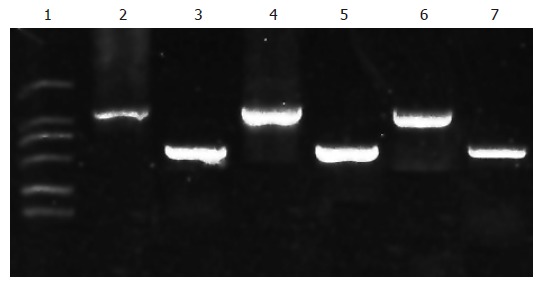
Electrophoregram of RT-PR product of annexin II . Lane 1: DNA marker DL 2000 (from top to bottom: 2000, 1000, 750, 500, 250, 100 bp); Lanes 2, 4 and 6: Annexin II from HUVEC, SMMC-7721 and HCC sample, respectively; Lanes 3, 5 and 7: β-actin RT-PCR product from HUVEC, SMMC-7721 and HCC sample, respectively.
Competitive inhibition
Using flow cytometry, we proved that β2GP I labeled with GFP could bind to SMMC-7721. The binding rate of β2GP I -GFP with SMMC-7721 (66.81%) was significantly higher than that of GFP with SMMC-7721 (1.16%) (Figure 4). Once β2GP I -GFP had been incubated with annexin II at room temperature in advance, the binding rate of β2GP I -GFP with SMMC-7721 dropped to 7.21% (Figure 4C). Thus these results suggested that annexin II could inhibit the combination of β2GP I with SMMC-7721.
Figure 4.
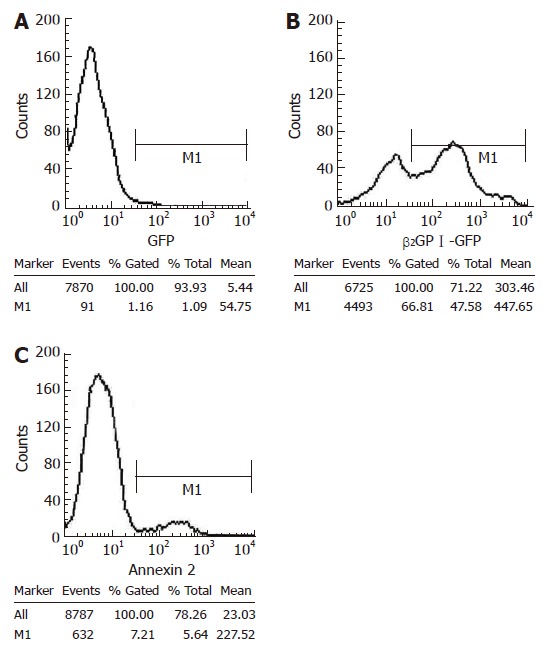
FACS analysis revealed descent of the binding rate of GFP-β2GP I with SMMC-7721 after pretreatment with annexin II .
DISCUSSION
Through cloning, recombining and sequencing the gene of β2GP I -bound receptor on membrane of SMMC-7721 cells, we found that the gene fragment of the receptor shared high homology with human annexin II (98%). Moreover, annexin II was found to exist on the membrane of SMMC-7721 by RT-PCR. At the same time, we validated that annexin II could compete with β2GP I for combining SMMC-7721. Thus, it can be concluded that the receptor specific for β2GP I might be annexin II .
Annexin II (Mr-36 ku) belongs to a family of Ca2+-dependent membrane-binding proteins encoded by some 20 different genes[10-12]. Annexins are structurally related proteins, each of which consists of an N-terminal “tail” and C-terminal “core” domain. The core domains of different annexins are highly conserved and share 40%-70% homology. Usually annexin II binds to S100A10 (p11), a Ca2+-modulated protein, to form tetramer. P11 can adulate the binding activity of annexin II for calcium ion or phospholipids. It has been shown that the gene of annexin II lies on the 15th chromosome, and has a 1.4-kb conserved encoding sequence. Despite the lack of a hydrophobic signal peptide, the presence of annexin II on cell surfaces is well established. It has been proven that annexin II is affluent on endotheliocyte, monocyte/macrophage, myeloid cell and some tumor cells. And approximately 4.3% of total endothelial annexin II is associated with the phospholipids on the external plasma membrane. Annexin II has been implicated to possess many biological functions in a variety of physiologic processes, such as anti-inflammatory effect of glucocorticoid, calcium-dependent exocytosis, immune response, calcium transport and phosphorlipase A2 regulation[12]. Recently, research has suggested that annexin II is an endothelial cell receptor for tissue-type plasminogen activator (t-PA) and plasminogen (PLG)[13-18] and can activate them. So under normal conditions, annexin II is an important modulation receptor in coagulation-anticoagulation-fibrinogenolysis system. Over-expression of annexin II will evoke hyperfunction of PLG and cause thrombosis and hemorrhage. As annexin II can act as second messenger in the modulation path of cell division, it has also been found to be related to cell proliferation and tumor growth. In addition, it has been suggested that the expression disturbance of annexin II in many kinds of cancers accelerate carcinogensis and metastasis.
With respect to the relationship between annexin II and virus infection, it has been elucidated that annexin II can serve as a receptor for cytomegalovirus and mediate its infection[19]. Until now, there lacks report on the relationship between annexin II and HBV infection. Our preliminary studies suggest that annexin II is the receptor of β2GP I , and β2GP I can bind to HBsAg. Therefore, we speculate that annexin II might have a potential role in HBV infection. To evaluate validity of the hypothesis and make sure which domain of annexin II binds to β2GP I so as to bridge HBV infection, we will use phage display in our further study.
Footnotes
Supported by The National Nature Science Foundation of China, No. 30070338
S- Editor Liu Y L- Editor Kumar M E- Editor Ma WH
References
- 1.Nimpf J, Bevers EM, Bomans PH, Till U, Wurm H, Kostner GM, Zwaal RF. Prothrombinase activity of human platelets is inhibited by beta 2-glycoprotein-I. Biochim Biophys Acta. 1986;884:142–149. doi: 10.1016/0304-4165(86)90237-0. [DOI] [PubMed] [Google Scholar]
- 2.Galli M, Comfurius P, Maassen C, Hemker HC, de Baets MH, van Breda-Vriesman PJ, Barbui T, Zwaal RF, Bevers EM. Anticardiolipin antibodies (ACA) directed not to cardiolipin but to a plasma protein cofactor. Lancet. 1990;335:1544–1547. doi: 10.1016/0140-6736(90)91374-j. [DOI] [PubMed] [Google Scholar]
- 3.McNeil HP, Simpson RJ, Chesterman CN, Krilis SA. Anti-phospholipid antibodies are directed against a complex antigen that includes a lipid-binding inhibitor of coagulation: beta 2-glycoprotein I (apolipoprotein H) Proc Natl Acad Sci USA. 1990;87:4120–4124. doi: 10.1073/pnas.87.11.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehdi H, Kaplan MJ, Anlar FY, Yang X, Bayer R, Sutherland K, Peeples ME. Hepatitis B virus surface antigen binds to apolipoprotein H. J Virol. 1994;68:2415–2424. doi: 10.1128/jvi.68.4.2415-2424.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehdi H, Yang X, Peeples ME. An altered form of apolipoprotein H binds hepatitis B virus surface antigen most efficiently. Virology. 1996;217:58–66. doi: 10.1006/viro.1996.0093. [DOI] [PubMed] [Google Scholar]
- 6.Stefas I, Rucheton M, D'Angeac AD, Morel-Baccard C, Seigneurin JM, Zarski JP, Martin M, Cerutti M, Bossy JP, Missé D, et al. Hepatitis B virus Dane particles bind to human plasma apolipoprotein H. Hepatology. 2001;33:207–217. doi: 10.1053/jhep.2001.20531. [DOI] [PubMed] [Google Scholar]
- 7.Gao P, Guo Y, Qu L, Shi T, Zhang H, Dong C, Yang H. [Relation between Beta-2-glycoprotein I and hepatitis B virus surface antigen] Zhonghua Gan Zang Bing Za Zhi. 2002;10:31–33. [PubMed] [Google Scholar]
- 8.Gao PJ, Piao YF, Liu XD, Qu LK, Shi Y, Wang XC, Yang HY. Studies on specific interaction of beta-2-glycoprotein I with HBsAg. World J Gastroenterol. 2003;9:2114–2116. doi: 10.3748/wjg.v9.i9.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao P, Piao Y, Wang X, Qu L, Shi Y, Yang H. A possible receptor for beta 2 glycoprotein I on the membrane of hepatoma cell line smmc7721. Chin Med J (Engl) 2003;116:1308–1311. [PubMed] [Google Scholar]
- 10.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 11.Moss SE, Morgan RO. The annexins. Genome Biol. 2004;5:219. doi: 10.1186/gb-2004-5-4-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raynal P, Pollard HB. Annexins: the problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim Biophys Acta. 1994;1197:63–93. doi: 10.1016/0304-4157(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Hajjar KA. Annexin II : a plasminogen-plasminogen activator co-receptor. Front Biosci. 2002;7:d341–d348. doi: 10.2741/kim. [DOI] [PubMed] [Google Scholar]
- 14.Hajjar KA, Guevara CA, Lev E, Dowling K, Chacko J. Interaction of the fibrinolytic receptor, annexin II , with the endothelial cell surface. Essential role of endonexin repeat 2. J Biol Chem. 1996;271:21652–21659. doi: 10.1074/jbc.271.35.21652. [DOI] [PubMed] [Google Scholar]
- 15.Hajjar KA, Jacovina AT, Chacko J. An endothelial cell receptor for plasminogen/tissue plasminogen activator. I. Identity with annexin II . J Biol Chem. 1994;269:21191–21197. [PubMed] [Google Scholar]
- 16.Hajjar KA, Mauri L, Jacovina AT, Zhong F, Mirza UA, Padovan JC, Chait BT. Tissue plasminogen activator binding to the annexin II tail domain. Direct modulation by homocysteine. J Biol Chem. 1998;273:9987–9993. doi: 10.1074/jbc.273.16.9987. [DOI] [PubMed] [Google Scholar]
- 17.Kassam G, Choi KS, Ghuman J, Kang HM, Fitzpatrick SL, Zackson T, Zackson S, Toba M, Shinomiya A, Waisman DM. The role of annexin II tetramer in the activation of plasminogen. J Biol Chem. 1998;273:4790–4799. doi: 10.1074/jbc.273.8.4790. [DOI] [PubMed] [Google Scholar]
- 18.Hajjar KA, Menell JS. Annexin II: a novel mediator of cell surface plasmin generation. Ann N Y Acad Sci. 1997;811:337–349. doi: 10.1111/j.1749-6632.1997.tb52013.x. [DOI] [PubMed] [Google Scholar]
- 19.Depla E. Interaction of viruses with annexins: a potential therapeutic target? Curr Opin Investig Drugs. 2000;1:415–420. [PubMed] [Google Scholar]


