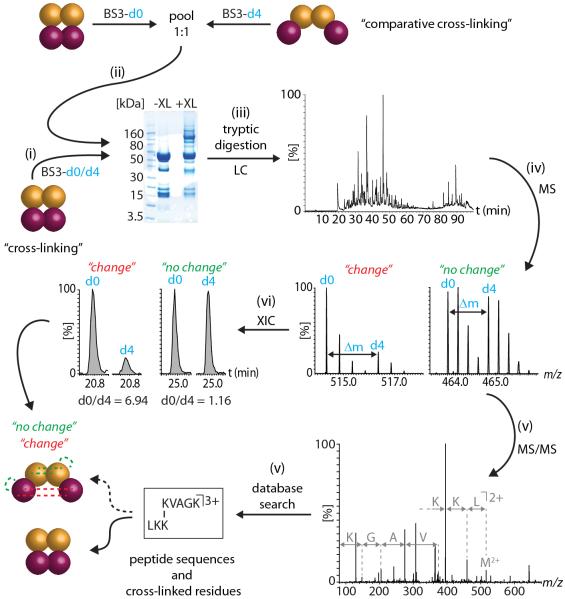
Figure 1. Comparative cross-linking workflow.
(i) The first step is to assign cross-links in the protein complex without perturbation or ligand binding. The protein complex is cross-linked using a 1:1 mixture of BS3-d0/d4. (ii) For comparative cross-linking, the same protein complex present in different conformations is cross-linked with non-deuterated (d0) and deuterated (d4) BS3, respectively. Both samples are subsequently pooled in equal quantities. Gel electrophoresis confirms cross-linking of the protein subunits by appearance of bands at higher molecular weight. (iii) The proteins are digested with trypsin and the peptide mixture containing cross-linked and non-cross-linked peptides is separated by nanoLC. (iv) Peptides are directly eluted into a mass spectrometer and peak pairs with a specific mass difference (Δm = 4Da) are indicative of light (d0) and heavy (d4) cross-linked peptides. (v) MS/MS and database searching identifies the cross-linked peptides. (vi) Ratios of the peptides corresponding to the different conformations of the protein complex (d0:d4) are determined by generation of extracted ion chromatograms (XICs) for both light (d0) and heavy (d4) cross-linked peptides.