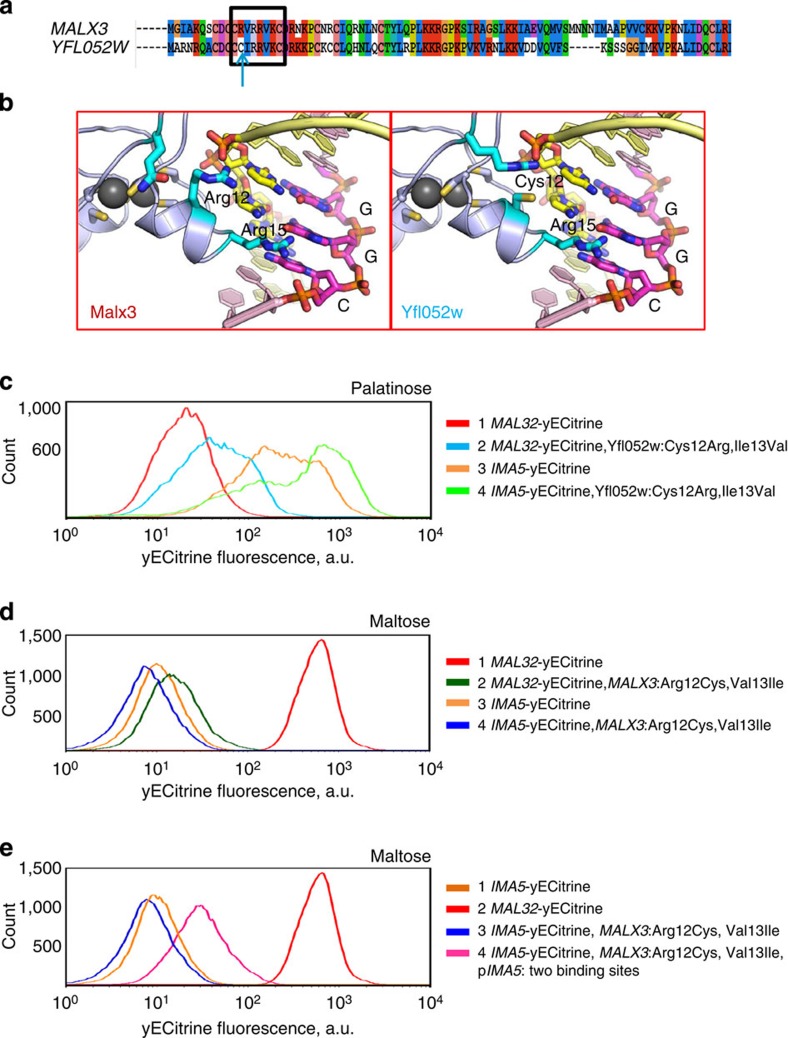
Figure 5. Differences in the DNA-binding domain of Malx3 and Yfl052w explain their different binding specificity.
(a) Alignment of the Malx3 and Yfl052w DNA-binding domains. Amino acids predicted to interact with the DNA-binding site are indicated with a black rectangle. The key position 12 that differs between Malx3 and Yfl052w is highlighted with a blue arrow. (b) Molecular modelling of the interaction between the Zn-finger domain and its DNA-binding site. Important base pairs are represented as yellow and magenta sticks, important amino acids are represented as blue sticks. The Arg15 is shared between both transcription factors and is responsible for the recognition of the G in the middle of the CGG binding motif. Arg12 in Malx3 does not take part in recognition of the CGG motif, but Cys12 in Yfl052w does interact with the DNA and is responsible for the preference for a G nucleotide in the third position of the motif. (c) A mutated version of the palatinose-specific Yfl052 activator (Cys12Arg and Ile13Val) is able to partly activate the MAL32 promoter in response to palatinose and also retains its capacity to activate the IMA5 promoter. (d) A mutated version of the maltose-specific Malx3 activator (Arg12Cys and Val13Ile) is incapable to activate MAL32 or IMA5 in response to maltose. (e) A mutated version of the maltose-specific Malx3 activator (Arg12Cys and Val13Ile) is capable to partly activate an IMA5 promoter containing an additional Yfl052w binding site. Each experiment was repeated at least three times with two biological replicates.