Figure 4.
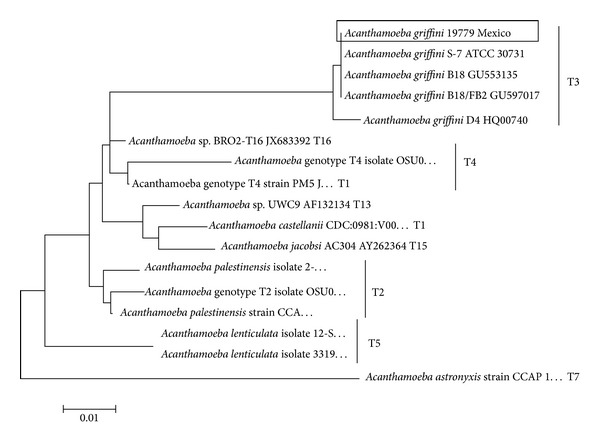
18S rRNA DF3 linearized Neighbour-Joining tree. The phylogenetic analysis was performed using the Kimura two-parameter distance algorithm in MEGA 5.0. The isolate obtained in the present study is identified in the tree (box). The type sequences were taken from GenBank and are presented under the following numbers: A. astronyxis strain CCAP 1534/1 genotype T7 Accession number AF239293, A. castellanii strain genotype T1 CDC:0981:V006 Accession number U07400, A. griffini S-7 ATCC 30731 genotype T3 Accession number U07412, A. griffini isolate B18 genotype T3 Accession number GU553135, A. griffini isolate B18/FB2 genotype T3 Accession number GU597017, A. griffini isolate D4 genotype T3 Accession number HQ00740, A. lenticulata isolate 12-SO #KC694184, A. lenticulata isolate 33195463 Accession number KC438381, Acanthamoeba sp. isolate OSU09-002 Accession number JQ669657, Acanthamoeba sp. genotype T2 Isolate OSU09-006 number JQ669661, Acanthamoeba palestinensis isolate TW-2 Accession number KC694193, A. palestinensis strain CCAP 1547-1 number AF239296, Acanthamoeba sp. isolate BRO2-T16 Accession number JX683392, Acanthamoeba sp. UWC9 genotype T13 Accession number AF132134, Acanthamoeba sp. PM5 genotype T4 Accession number JX494395, Acanthamoeba sp. genotype T4 Accession number JQ669660, and Acanthamoeba jacobsi AC304 Accession number AY262364.
