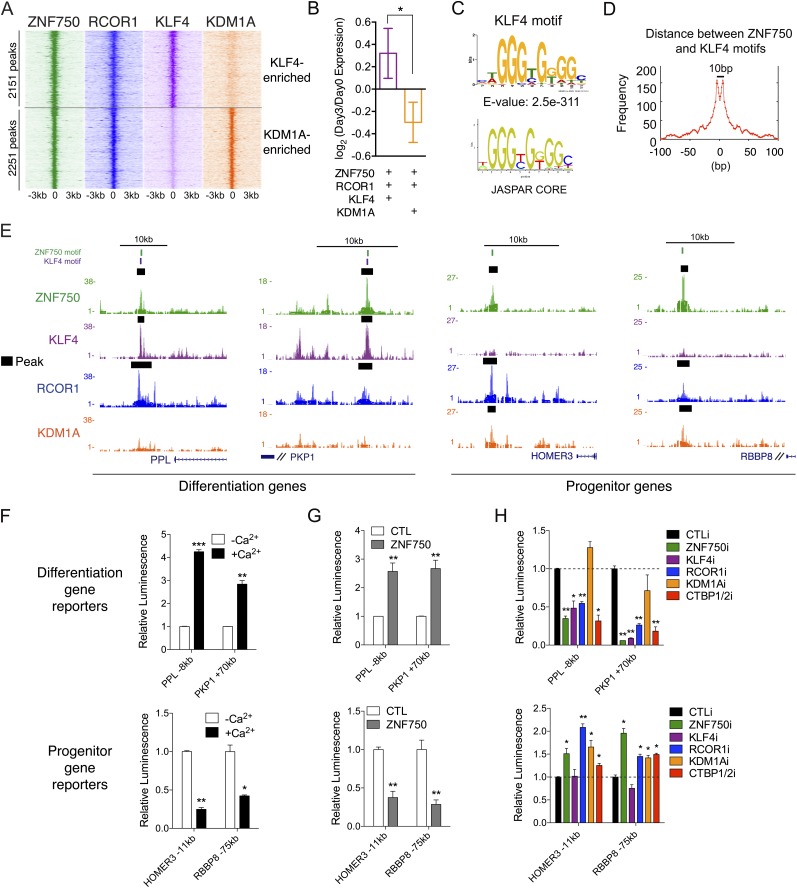
Figure 5.
ZNF750 activates or represses transcription with different interacting proteins. (A) Heat map of combined ZNF750, KLF4, RCOR1, and KDM1A ChIP-seq peaks, selected for KLF4 or KDM1A enrichment in ZNF750- and RCOR1-binding regions. Heat map shows ChIP-seq signal over 6-kb regions around peak center. (B) Log2 fold change during keratinocyte differentiation of genes near the top 500 ZNF750, KLF4, and RCOR1 peaks or the ZNF750, RCOR1, and KDM1A peaks. (*) P < 0.05. (C) Sequence-specific motif identified in KLF4 ChIP-seq peaks, compared with canonical KLF4 motif. (D) Frequency of distances between pairs of ZNF750 and KLF4 motifs in ChIP-seq peaks. (E) Example ZNF750, KLF4, RCOR1, and KDM1A ChIP-seq tracks near two differentiation genes (left) and two progenitor genes (right), showing locations of motifs for ZNF750 (green) and KLF4 (purple). (Black bars) MACS-called peaks. (F) Change in luciferase activity of reporters near differentiation genes (top) or progenitor genes (bottom) during calcium-induced keratinocyte differentiation. t-test: (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, compared with –Ca2+. (G) Change in luciferase activity of each reporter with ectopic ZNF750 expression in progenitor keratinocytes compared with empty vector control (CTL). (**) P < 0.01, compared with CTL. (H) Luciferase activity of reporters with control knockdown (CTLi) compared with depletion of ZNF750, KLF4, RCOR1, KDM1A, or CTBP1/2. (*) P < 0.05; (**) P < 0.01, compared with CTLi. See also Supplemental Figure S5 and Supplemental Tables S2–S6.