Abstract
Objective
This work focuses on multiply-related Unified Medical Language System (UMLS) concepts, that is, concepts associated through multiple relations. The relations involved in such situations are audited to determine whether they are provided by source vocabularies or result from the integration of these vocabularies within the UMLS.
Methods
We study the compatibility of the multiple relations which associate the concepts under investigation and try to explain the reason why they co-occur. Towards this end, we analyze the relations both at the concept and term levels. In addition, we randomly select 288 concepts associated through contradictory relations and manually analyze them.
Results
At the UMLS scale, only 0.7% of combinations of relations are contradictory, while homogeneous combinations are observed in one-third of situations. At the scale of source vocabularies, one-third do not contain more than one relation between the concepts under investigation. Among the remaining source vocabularies, seven of them mainly present multiple non-homogeneous relations between terms. Analysis at the term level also shows that only in a quarter of cases are the source vocabularies responsible for the presence of multiply-related concepts in the UMLS. These results are available at: http://www.isped.u-bordeaux2.fr/ArticleJAMIA/results_multiply_related_concepts.aspx.
Discussion
Manual analysis was useful to explain the conceptualization difference in relations between terms across source vocabularies. The exploitation of source relations was helpful for understanding why some source vocabularies describe multiple relations between a given pair of terms.
Keywords: Artificial Intelligence, Systems Integration, Terminology, Unified Medical Language System, Terminology auditing
Introduction
Decades of natural language processing and artificial intelligence research on the methods for terminology acquisition and structuring1 have resulted in an increasing number of available terminologies. These terminologies often describe complementary features of scientific and technical areas. Consequently, their integration can be useful for the comprehensive description and modeling of these areas. Moreover, the issues specific to integration may also be important in other contexts, such as maintenance, updating,2 evolution,3 4 and transcoding or alignment5 of terminologies and ontologies. As an illustration, the Unified Medical Language System (UMLS)6 7 integrates over 170 biomedical terminologies. The result of this integration is particularly useful and widely employed in the biomedical area for various applications (eg, information retrieval and extraction, coding of discharge patient records, question/answering systems).
Although sometimes necessary, integration may however also cause conceptual and structural inconsistencies. At the scale of a single terminology, inconsistencies may be found, while the situation becomes more complex when terminologies are merged. Indeed, terminologies are designed and created with different objectives, and have different underlying principles. In order to detect and correct these potential limitations, researchers have proposed methods for auditing terminological resources. Such methods have been applied to WordNet8 for redundancy and consistency checking.9–12 In order to apply this approach to the biomedical domain, we adopt the analysis grid proposed in a recent review of auditing methods,13 and distinguish three aspects:
Terms and concepts: this aspect focuses on term labeling,14 ambiguity and polysemy,15–17 synonymy completeness,18–22 coverage for a given subdomain or application,18 21 23–26 and modifier influence.27
Semantic categorization: the consistency of the UMLS semantic categorization of concepts is checked according to the hierarchical relations associating these concepts.16 26 28–33
Semantic relationships: the consistency of hierarchical relationships and their coverage have been widely studied,15 19 23 28 30 34–41 while other types of relationships have not been studied properly to date.
Our study concerns the first and third aspects. We propose analyzing the multiply-related UMLS concepts, that is, concepts which are associated through multiple relationships. This situation arises within the UMLS because, during the integration of source vocabularies, any information related to terms and relations is preserved. In our opinion, beyond the integration of synonymous terms and the increase in the lexical coverage provided by a single terminology, the generation of multiply-related concepts is another artifact of terminology integration. To our knowledge, this aspect has not yet been systematically investigated. The closest work,42 which focuses on such UMLS concepts, only performs manual categorization and review of some common situations. In our study, we propose automatic methods to audit all multiply-related UMLS concepts. Basically, we study the compatibility of the multiple relations which associate the concepts under investigation and seek to explain the reason why they co-occur. In addition, we randomly select 288 concepts associated through contradictory relations and manually analyze them. Our previous work43 is strengthened with several detailed analyses of the data. In addition, new aspects have been added, such as clarification of the main terms used in the paper and investigation of incompatibilities between relations at the scale of source terminologies, especially by considering the term level.
Material
The UMLS is a terminological system whose main component, the Metathesaurus, integrates 173 source vocabularies and represents a huge graph composed of 2 669 792 concepts. A concept corresponds to a set of synonymous terms provided by different source vocabularies (in this paper, we use ‘source vocabularies’ and ‘source terminologies’ interchangeably). The concepts are organized within a very dense terminological network: 53 942 132 binary relations linking concepts are recorded in the investigated version of the UMLS (2012AA). Such a huge quantity of relations is due to the fact that, according to the UMLS building rules, all the relations existing in the source vocabularies have to be integrated within the UMLS, even when there are conflicting relations between two concepts.
There are 11 types of active UMLS relationships in the 2012AA version, which can be grouped into three general classes (table 1):
Synonymy
Hierarchical
Associative.
Table 1.
UMLS relationships and the class to which they belong
| Class of relationship | Abbreviation of the relationship | Meaning of the relationship |
|---|---|---|
| Synonymy | SY | Source asserted synonymy |
| Hierarchical | CHD | Has child relationship |
| PAR | Has parent relationship | |
| RB | Has a broader relationship | |
| RN | Has a narrower relationship | |
| SIB | Has sibling relationship | |
| Associative | AQ | Allowed qualifier |
| QB | Can be qualified by | |
| RO | Has relationship other than synonymous, narrower or broader | |
| RL | Has similar or ‘alike’ relationship | |
| RQ | Related and possibly synonymous |
Regarding the relationships at the scale of source vocabularies, the Metathesaurus records around 300 source relationships and assigns each of them to one of the 11 active UMLS relationships according to the source vocabularies documentation or their interpretation by the National Library of Medicine (NLM) team. For instance, the source relationship same_as is assigned to the SY relationship, inverse_isa to PAR, and has_component to RO.
Methods
For a given pair of concepts (C1, C2) associated through multiple relations, we study the compatibility between relationships at the UMLS scale and at the scale of source vocabularies. We then determine the reason for the presence of multiple relations between concept pairs. Before presentation of the methods, we define the main terms used and explain the material preparation.
Definitions
Relationship and relation: while relationship indicates a given type of relation (eg, synonymy relationship, hierarchical relationship), relation refers to every individual link between two given terms.
Multiply-related concepts: Two distinct UMLS concepts that are associated through at least two relationships, are multiply-related concepts. For example, Butyrolactone (C0178525) is multiply related to Lactones (C0022947) through PAR and RB.
Source relation(ship): A source relation(ship) corresponds to a relation(ship) which comes from source terminologies distinct from the 11 active UMLS relationships.
Symmetric relationships: Symmetric relationships correspond to relationships which can be read identically in both senses, that is, if a triplet (C1, R, C2) exists, then the triplet (C2, R, C1) is also present. Among the UMLS relationships, RL, RO, RQ, SIB and SY are symmetric.
Inverse relationships: When two concepts are linked to each other through reciprocal relationships from the same class, these relationships are characterized as inverse. The inverse relationships present in the UMLS are AQ/QB, PAR/CHD and RB/RN.
Material preparation and de-duplication
As mentioned previously, all (source) relations existing in the source vocabularies are integrated within the UMLS. Consequently, some relations may be redundant when described by distinct vocabularies. This results in identical triplets, of which we keep only one specimen. For example, the SY relation existing between Adrenocortical hyperfunction (C0001622) and Cushing's syndrome (C0010481) is defined both in MEDCIN and SNOMEDCT. Only one triplet (Adrenocortical hyperfunction, SY, Cushing's syndrome) is considered here.
Due to other building principles, binary relations are represented in both directions within the UMLS. In practice, if a source vocabulary describes that a given asymmetric/symmetric relation exists between C1 and C2, then its inverse/the same relation is also recorded between C2 and C1 when the source vocabulary is integrated within the UMLS. We de-duplicate such situations so that we do not analyze the same triplet twice. For instance, of the two triplets (Butyrolactone, RB, Lactones) and (Lactones, RN, Butyrolactone), only the first one is kept.
Finally, the labeled relationships mapped_to and mapped_from are defined as ‘one-to-one mappings between two vocabularies which are both present in the UMLS.’ We thus choose to ignore them because they are generated by the UMLS based on maps between source terminologies, and so are different from other relationships.
Compatibility of relationships associating multiply-related concepts
We study the compatibility of the relationships which associate multiply-related concepts and distinguish four situations (figure 1):
Contradictory combinations: combinations which include inverse relationships (eg, CHD PAR, RB RL RN, AQ QB, PAR RN RO SIB). Although not natively inverse, the combinations involving PAR with RN and RB with CHD are also considered as contradictory
Granularity difference: combinations which include SIB and/or SY combined with at least one of the following hierarchical relationships: PAR, CHD, RB, RN (eg, PAR SIB, PAR RO SIB, RB SY)
Heterogeneous combinations: combinations involving relationships from distinct classes (ie, synonymy, hierarchical, associative) (eg, PAR RO, RQ SIB, PAR RB RO RQ)
Homogeneous combinations: the two combinations PAR RB and CHD RN in addition to any combination of relationships within the associative class except for AQ QB because they are inverse (eg, RO RL, AQ RO RQ).
Figure 1.
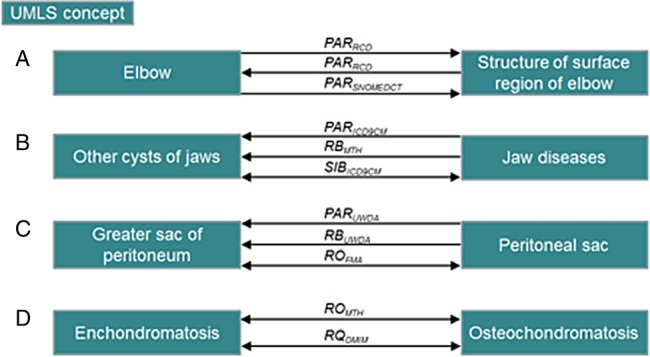
The different categories of relation combinations between multiply-related concepts: (A) contradictory combinations, (B) combinations with granularity difference, (C) heterogeneous combinations, (D) homogeneous combinations.
The pairs are associated with only one of these categories in the order of their presentation. Thus, if a concept pair exhibits both contradictory and homogeneous combinations, then the pair is only counted in contradictory combinations.
The compatibility of relationships is investigated at the UMLS scale and at the scale of source terminologies. In addition, since contradictory combinations produce the most problematic situations, we manually analyze 10% of concept pairs randomly extracted from this set.
Reason for the presence of multiple relations between concepts
We want to determine, for each pair of concepts under investigation, if its combination of relations already exists in source vocabularies or if it results from their integration within the UMLS. Towards this end, for each pair of concepts, we first check if at least one source terminology describes its combination. If not, then the presence of multiple relations is due to the UMLS integration process. If the combination is observed in a given source vocabulary, it is necessary to check if it also exists at the term level. In practice, we analyze the terms which are clustered into multiply-related concepts and consider that the combination actually originates from the source vocabulary if the same pair of terms is multiply-related (figure 2).
Figure 2.
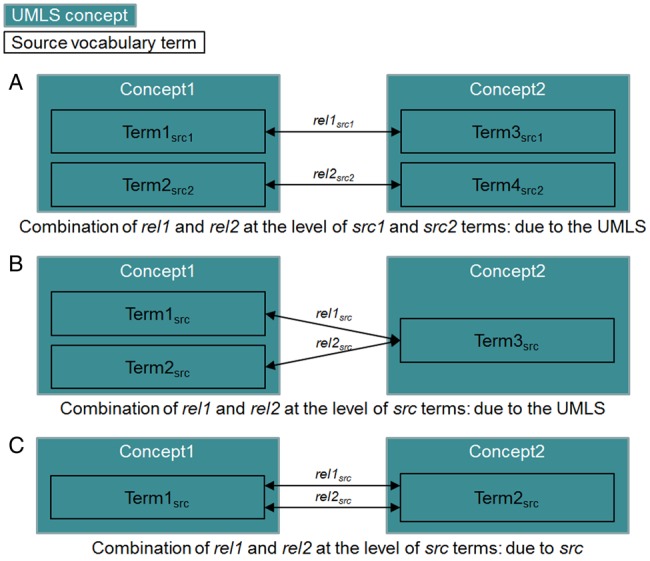
Analysis of multiple relations between UMLS concepts at the term level: (A) and (B) combinations generated during the UMLS integration process, (C) combinations already present in a source vocabulary.
Results
After de-duplication, the number of distinct concept pairs is 12 356 156 (involving 2 669 792 distinct concepts). Our study addresses the 439 087 concept pairs (involving 360 098 distinct concepts) which are associated through multiple relations, corresponding to 3.6% of the total number of concept pairs related within the UMLS (involving 13.5% of distinct concepts). The results presented in this section are available at: http://www.isped.u-bordeaux2.fr/ArticleJAMIA/results_multiply_related_concepts.aspx.
Compatibility of relationships associating multiply-related concepts
At the UMLS scale (table 2), combinations exhibiting contradictions and granularity differences represent 0.7% and 20.0% of the investigated concept pairs, respectively. Heterogeneous combinations are the most frequent (45.4%) and homogeneous combinations represent 33.9% of all investigated concept pairs. A total of 157 combinations are observed, and the 10 most common cover over 88.0% of the entire set. The three most frequent combinations correspond to different situations: homogeneous PAR RB (31.4%), heterogeneous PAR RO (26.4%) and granularity difference PAR SIB (11.6%). Conversely, 60 combinations (eg, CHD PAR RB RL RN RQ SIB, PAR RN SY, AQ RB, RB RL RO SY) occur less than 10 times. Although only 0.7% of concept pairs are related through contradictory combinations, this situation shows the highest number of combinations (59.9%).
Table 2.
Compatibility of relationships associating multiply-related concepts at the UMLS scale
| Contradictory combinations | Granularity difference | Heterogeneous combinations | Homogeneous combinations | Total | |
|---|---|---|---|---|---|
| No. of multiply-related concept pairs | 2880 (0.7%) | 88038 (20.0%) | 199272 (45.4%) | 148 897 (33.9%) | 439 087 (100.0%) |
| No. of combinations | 94 (59.9%) | 36 (22.9%) | 23 (14.7%) | 4 (2.5%) | 157 (100.0%) |
| Most frequent combinations | CHD PAR (628) PAR RN (472) RB RN (159) | PAR SIB (50 785) PAR RB SIB (11 924) RB SIB (6951) | PAR RO (115 757) PAR RB RO (25 055) RB RO (18 281) | PAR RB (137 918) RO RQ (10 916) AQ RO (61) |
At the scale of source vocabularies, the concepts investigated are provided by 66 source terminologies (table 3). We distinguish two categories:
Twenty-two source vocabularies do not exhibit any combination of relationships (eg, HLREL, KCD5, MMSL, MTH, NIC, RXNORM). In other words, these source terminologies always contain only one relationship between two concepts.
Forty-four source vocabularies describe multiple relations between the concepts under investigation (table 4). Only seven of them (all containing fewer than 70 pairs of multiply-related concepts) present a high percentage of contradictory combinations (eg, 100% for ICD10, CST and DSM4). Eleven source terminologies contain mainly concepts multiply related through relations with granularity difference (eg, over 97% for RCD, ICD9CM and ICD10AM), while 15 source vocabularies mainly exhibit heterogeneous combinations (eg, over 99% for LNC, NDFRT and NCI). The 11 remaining source terminologies exhibit a high percentage of concept pairs related through homogeneous combinations (eg, over 95% for AOD, CSP, PSY and NEU).
Table 3.
Acronym and name of source vocabularies providing multiply-related concepts
| Vocabulary acronym | Vocabulary name |
|---|---|
| AIR | AI/RHEUM |
| ALT | Alternative Billing Concepts |
| AOD | Alcohol and Other Drug Thesaurus |
| AOT | Authorized Osteopathic Thesaurus |
| BI | Beth Israel Vocabulary |
| CCC | Clinical Care Classification |
| CCPSS | Canonical Clinical Problem Statement System |
| CCS | Clinical Classifications Software |
| CPM | Medical Entities Dictionary |
| CPT | Physicians’ Current Procedural Terminology |
| CSP | CRISP Thesaurus |
| CST | COSTART |
| DSM3R | DSM-III-R |
| DSM4 | DSM-IV |
| FMA | Foundational Model of Anatomy Ontology |
| GO | Gene Ontology |
| HCPCS | Metathesaurus HCPCS Hierarchical Terms |
| HHC | Home Health Care Classification |
| HL7V2.5 | HL7 Vocabulary V.2.5 |
| HL7V3.0 | HL7 Vocabulary V.3.0 |
| ICD10 | ICD10 |
| ICD10AM | International Statistical Classification of Diseases and Related Health Problems |
| ICD10CM | International Classification of Diseases |
| ICD10PCS | ICD-10-PCS |
| ICD9CM | ICD-9-CM |
| ICF | International Classification of Functioning |
| ICNP | International Classification for Nursing Practice |
| ICPC | ICPC |
| ICPC2P | ICPC-2 PLUS |
| JABL | Online Congenital Multiple Anomaly/Mental Retardation Syndromes |
| KCD5 | Korean Standard Classification of Disease V.5 |
| LNC | LOINC 2.15 |
| MDR | Medical Dictionary for Regulatory Activities Terminology (MedDRA) |
| MEDCIN | MEDCIN |
| MEDLINEPLUS | MedlinePlus Health Topics |
| MMSL | Multum MediSource Lexicon |
| MSH | Medical Subject Headings |
| MTH | UMLS Metathesaurus |
| MTHMST | Metathesaurus Version of Minimal Standard Terminology Digestive Endoscopy |
| MTHSPL | Metathesaurus FDA Structured Product Labels |
| NAN | NANDA Nursing Diagnoses: Definitions and Classification |
| NCBI | NCBI Taxonomy |
| NCI | NCI Thesaurus |
| NDFRT | National Drug File |
| NEU | Neuronames Brain Hierarchy |
| NIC | Nursing Interventions Classification (NIC) |
| NOC | Nursing Outcomes Classification |
| OMIM | Online Mendelian Inheritance in Man |
| OMS | Omaha System |
| PCDS | Patient Care Data Set |
| PDQ | Physician Data Query |
| PNDS | Perioperative Nursing Data Set |
| PPAC | Pharmacy Practice Activity Classification |
| PSY | Thesaurus of Psychological Index Terms |
| RAM | QMR clinically related terms from Randolph A. Miller |
| RCD | Read Thesaurus |
| RXNORM | RxNorm Vocabulary |
| SNM | SNOMED-2 |
| SNMI | SNOMED International |
| SNOMEDCT | SNOMED Clinical Terms |
| ULT | UltraSTAR |
| UMD | UMDNS: Product Categories Thesaurus |
| USPMG | USP Model Guidelines |
| UWDA | University of Washington Digital Anatomist |
| VANDF | Veterans Health Administration National Drug File |
| WHO | WHOART |
Table 4.
Compatibility of relationships associating multiply-related concepts at the scale of source vocabularies
| Vocabulary acronym | Contradictory combinations | Granularity difference |
Heterogeneous combinations | Homogeneous combinations | Total | Pairs uniquely related at the term level |
Pairs multiply related at the term level |
Number of combinations | Most frequent combination |
|---|---|---|---|---|---|---|---|---|---|
| UWDA | 2 (0.0%) | 2474 (2.8%) | 6439 (7.4%) | 78 568 (89.8%) | 87 483 | 406 (0.5%) | 87 077 (99.5%) | 9 | PAR RB (89.8%) |
| LNC | 5 (0.0%) | 64 948 (100.0%) | 64 953 | 64 953 (100.0%) | 2 | PAR RO (100.0%) | |||
| RCD | 241 (0.7%) | 34 855 (99.3%) | 35 096 | 34 920 (99.5%) | 176 (0.5%) | 4 | PAR SIB (98.8%) | ||
| SNOMEDCT | 242 (1.6%) | 343 (2.3%) | 14 021 (92.1%) | 615 (4.0%) | 15 221 | 15 036 (98.8%) | 185 (1.2%) | 29 | PAR RO (86.7%) |
| MEDCIN | 49 (0.3%) | 209 (1.4%) | 1919 (13.1%) | 12 505 (85.2%) | 14 682 | 14 678 (100.0%) | 4 (0.0%) | 16 | PAR RB (85.1%) |
| AOD | 12 (0.1%) | 317 (2.3%) | 299 (2.2%) | 13 209 (95.5%) | 13 837 | 345 (2.5%) | 13 492 (97.5%) | 27 | PAR RB (95.1%) |
| CSP | 82 (0.7%) | 412 (3.5%) | 11 245 (95.8%) | 11 739 | 166 (1.4%) | 11 573 (98.6%) | 13 | PAR RB (95.4%) | |
| NDFRT | 11 (0.1%) | 9393 (99.9%) | 9404 | 7874 (83.7%) | 1530 (16.3%) | 3 | PAR RO (99.9%) | ||
| ICD10CM | 76 (0.9%) | 7550 (94.2%) | 388 (4.8%) | 8014 | 8014 (100.0%) | 9 | PAR SIB (93.7%) | ||
| ICD9CM | 37 (0.6%) | 5979 (97.3%) | 129 (2.1%) | 6145 | 6145 (100.0%) | 10 | PAR SIB (96.5%) | ||
| ICD10AM | 73 (1.2%) | 5930 (98.8%) | 2 (0.0%) | 6005 | 6005 (100.0%) | 6 | PAR SIB (98.2%) | ||
| PSY | 19 (0.3%) | 5548 (99.7%) | 5567 | 27 (0.5%) | 5540 (99.5%) | 7 | PAR RB (99.3%) | ||
| MDR | 69 (1.6%) | 4136 (93.4%) | 221 (5.0%) | 4426 | 4426 (100.0%) | 11 | PAR SIB (92.8%) | ||
| GO | 15 (0.5%) | 1224 (37.9%) | 553 (17.1%) | 1438 (44.5%) | 3230 | 1301 (40.3%) | 1929 (59.7%) | 33 | RB SIB (32.4%) |
| WHO | 1018 (35.8%) | 32 (1.1%) | 1790 (63.0%) | 2840 | 585 (20.6%) | 2255 (79.4%) | 11 | PAR RB (63.0%) | |
| NCI | 8 (0.3%) | 1 (0.0%) | 2694 (99.7%) | 2703 | 71 (2.6%) | 2632 (97.4%) | 5 | PAR RO (99.4%) | |
| FMA | 1 (0.0%) | 196 (9.3%) | 1901 (90.6%) | 2098 | 536 (25.5%) | 1562 (74.5%) | 5 | RO SIB (79.2%) | |
| MEDLINEPLUS | 119 (6.1%) | 1828 (93.8%) | 1 (0.1%) | 1948 | 373 (19.1%) | 1575 (80.9%) | 10 | RO SIB (82.2%) | |
| MSH | 8 (0.4%) | 760 (40.0%) | 1109 (58.4%) | 22 (1.2%) | 1899 | 960 (50.6%) | 939 (49.4%) | 11 | PAR SIB (38.1%) |
| NEU | 1 (0.1%) | 805 (99.9%) | 806 | 1 (0.1%) | 805 (99.9%) | 2 | PAR RB (99.9%) | ||
| UMD | 11 (1.5%) | 102 (13.8%) | 599 (81.3%) | 25 (3.4%) | 737 | 234 (31.8%) | 503 (68.2%) | 17 | RO SIB (54.8%) |
| VANDF | 588 (100.0%) | 588 | 588 (100.0%) | 1 | RB RO (100.0%) | ||||
| SNMI | 135 (29.5%) | 322 (70.5%) | 457 | 457 (100.0%) | 3 | PAR RO (70.2%) | |||
| OMS | 376 (100.0%) | 376 | 376 (100.0%) | 1 | PAR RO (100.0%) | ||||
| AOT | 6 (1.7%) | 91 (25.6%) | 258 (72.7%) | 355 | 2 (0.6%) | 353 (99.4%) | 3 | PAR RB (72.7%) | |
| ICF | 263 (100.0%) | 263 | 263 (100.0%) | 1 | PAR SIB (100.0%) | ||||
| CCS | 1 (0.4%) | 47 (19.0%) | 200 (80.6%) | 248 | 248 (100.0%) | 8 | PAR RQ (75.4%) | ||
| PDQ | 4 (2.1%) | 7 (3.7%) | 177 (94.1%) | 188 | 11 (5.9%) | 177 (94.1%) | 4 | PAR RO (94.1%) | |
| NOC | 2 (2.1%) | 93 (97.9%) | 95 | 95 (100.0%) | 2 | PAR SIB (97.9%) | |||
| ICD10 | 61 (100.0%) | 61 | 61 (100.0%) | 1 | CHD PAR (100.0%) | ||||
| SNM | 18 (62.1%) | 11 (37.9%) | 29 | 29 (100.0%) | 2 | CHD PAR (62.1%) | |||
| CST | 21 (100.0%) | 21 | 21 (100.0%) | 1 | CHD PAR (100.0%) | ||||
| OMIM | 20 (100.0%) | 20 | 20 (100.0%) | 1 | RO RQ (100.0%) | ||||
| CPM | 14 (100.0%) | 14 | 14 (100.0%) | 1 | PAR SIB (100.0%) | ||||
| DSM4 | 11 (100.0%) | 11 | 11 (100.0%) | 1 | CHD PAR (100.0%) | ||||
| CPT | 10 (100.0%) | 10 | 10 (100.0%) | 1 | CHD PAR (100.0%) | ||||
| HHC | 8 (100.0%) | 8 | 8 (100.0%) | 1 | PAR SIB (100.0%) | ||||
| DSM3R | 6 (100.0%) | 6 | 6 (100.0%) | 1 | CHD PAR (100.0%) | ||||
| ALT | 6 (100.0%) | 6 | 6 (100.0%) | 1 | PAR SIB (100.0%) | ||||
| USPMG | 4 (100.0%) | 4 | 4 (100.0%) | 1 | PAR SIB (100.0%) | ||||
| HL7V3.0 | 2 (100.0%) | 2 | 2 (100.0%) | 1 | PAR RO (100.0%) | ||||
| MTHMST | 1 (100.0%) | 1 | 1 (100.0%) | 1 | RB RO (100.0%) | ||||
| BI | 1 (100.0%) | 1 | 1 (100.0%) | 1 | RO RQ (100.0%) | ||||
| NCBI | 1 (100.0%) | 1 | 1 (100.0%) | 1 | CHD PAR (100.0%) |
Except for the last two columns, the indicated numbers correspond to pairs of concepts which are associated through combinations of relations or through unique/multiple relations at the term level.
Source vocabularies highlighted in italics mainly present multiple relations between terms whose combination is homogeneous, while those highlighted in bold mainly present multiple relations between terms whose combination is not homogeneous.
Reason for the presence of multiple relations between concepts
For 179 963 concept pairs (41.0%), the combination of relations is not present in source terminologies and thus appears during their integration into the UMLS. For example, Skeletal system (C0037253) is related to Skeletal bone (C0262950) through CHD in LNC (represented by a PAR relationship from Skeletal system to Bones in figure 3A) and PAR in RCD. When investigating the term level for the remaining 259 124 concept pairs, we discover that for 150 109 concept pairs (34.2%), distinct term pairs are associated through unique relations. This means that the presence of multiple relations between the corresponding concepts actually results from the UMLS integration process, when the different terms were clustered into a unique concept. As an illustration, Ameboma of intestine (C0494031) is related to Amebiasis (C0002438) through PAR and SIB according to ICD10CM. However, the ICD10CM term belonging to the former concept, Ameboma of intestine, is associated with the ICD10CM terms Amebiasis and Amebiasis, unspecified (part of the second concept) through PAR and SIB, respectively (figure 3B). Consequently, ‘only’ 109 015 concept pairs (24.8%) are already multiply related in source vocabularies. For example, DNA, A-form (C0000702) is related to Nucleic acid conformation (C0028599) through PAR and RO because the corresponding MSH terms are associated through these multiple relations (figure 3C).
Figure 3.
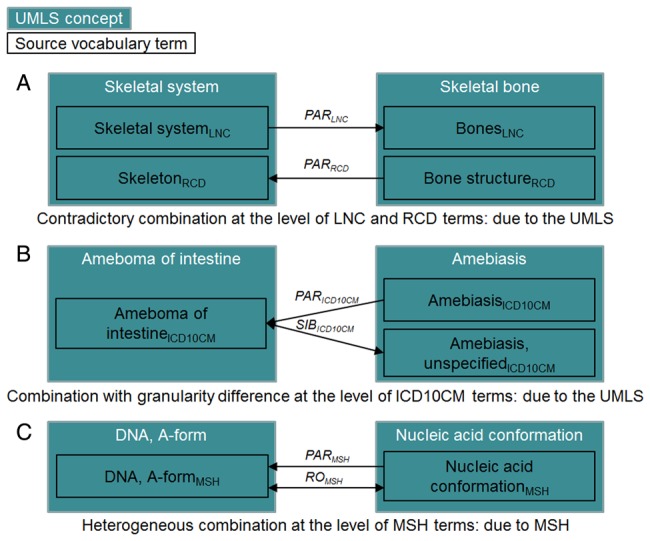
Illustrations of multiple relations between UMLS concepts at the term level: (A) a contradictory combination generated during the UMLS integration process, (B) a combination exhibiting granularity difference generated during the UMLS integration process, (C) a heterogeneous combination already present in MSH.
At the scale of source terminologies, we further analyze the 44 source terminologies exhibiting multiple relations between the concepts under investigation. We indicate in the seventh and eighth columns of table 4 whether the relations are unique or multiple at the term level. The 14 source terminologies exhibiting mainly pairs multiply related at the term level are those which require further investigation because they associate a given pair of terms through multiple relations. Among them, two profiles can be distinguished:
Seven source vocabularies mainly associate terms through homogeneous combinations of relations (eg, UWDA, AOD, CSP and PSY), PAR RB most of the time (highlighted in italics in table 4).
The seven remaining source terminologies principally relate terms with non-homogeneous combinations (eg, GO, NCI, FMA and MEDLINEPLUS), such as RB SIB, PAR RO and RO SIB (highlighted in bold in table 4).
Detailed analysis of contradictory combinations
We first determine the reason for the presence of multiple relations between the 288 pairs of concepts under investigation (10% of 2880). For 182 of them, the combination of relations does not exist in the source vocabularies. For the 106 remaining pairs, only three of them are actually multiply related in source terminologies. For example, Common bile duct (C0009437) and Hepatopancreatic ampulla (C0042425) are related through CHD, PAR, RN and RO in UWDA. Thus, UMLS integration is responsible for 99.9% of these contradictory combinations.
Manual analysis of these 288 pairs indicates that contradictory combinations often exist because of inherent or terminology-induced semantic features of terms:
The semantic value of compounds which coordinate terms (colorectal and colon/rectum), as previously observed in Bodenreider et al,27 and of the coordination (and/or) in general may differ according to source vocabularies. For instance, in MDR, Esophageal stenosis (C0014866) is parent of Oesophageal stenosis and obstruction (C0851721), while the relation is inverted in MEDCIN. We assume this situation is due to the meaning given to the coordination: it may be used to create more general terms or to specify terms. The semantic meaning of the unspecified (NOS, NEC, etc) modifier can also vary according to source terminologies.
The implicit nature of some modifiers may have an impact on semantic relations. For example, Total nephrectomy (C0176996) and Nephrectomy (C0027695) are associated through five relations (PAR, RN, RO, SIB, SY). The difference seems to be due to the total modifier, which may be implicit (SY is then proposed) or not (other relationships are then proposed).
Functional and causal links between terms also present great variations when they are transformed into hierarchical relationships. For instance, Angioedema (C0002994) and Urticaria (C0042109), which are common manifestations of allergic reactions, are related through CHD, PAR, RB, RO, RQ and SIB.
The ambiguity of some components of the terms can result in contradictory relationships across the source vocabularies. For example, Adrenal gland diseases (C0001621) and Dysfunction adrenal (C0549609) are associated through PAR, RN and RQ, in which dysfunction can mean (broadly) any disorder, or (narrowly) specific conditions in which endocrine function is either increased or decreased.
The difficulty for reflecting the structure of chemical products sometimes results in contradictory relations representing the different compounds. For instance, the link between Clodronic acid (C0012081) and Clodronate (C0162357) attempts to translate the chemical derivation of products into hierarchical (PAR, RB, RN) or other (RO) relationships.
- Terms with descriptive labels or terms reflecting complex biomedical notions, coupled with the choice of source vocabularies to create all possible links between such terms, may lead to inconsistencies. In fact, this situation may involve and accentuate any other cause discussed above:
- The meaning of terms may overlap, while each of them may have its own modifier(s), such as skin and other in the pair Skin diseases, bullous (C0085932) and Other bullous disorders (C0494828)
- They may combine several other causes already mentioned.
Discussion
Findings and limitations
Our study has interesting findings. At the UMLS scale, contradictory combinations are infrequent and may result from the fact that the conceptualization of relations between terms can be very different across source terminologies. Explanations provided by manual analysis cast some light on this difference. Homogeneous combinations are observed in one-third of situations, indicating the presence of redundant relations between concepts. At the scale of source terminologies, compatibility analysis reveals that one-third of them always describe only one relation between a given pair of concepts. Among the 44 remaining source vocabularies, 14 of them use multiple relations to associate a unique pair of terms. When the combination of these multiple relations is homogeneous, this indicates distinct but coherent points of view for expressing a link between two terms. On the other hand, when the relations constitute non-homogeneous combinations, the situation is more troublesome because this indicates that a given source terminology describes multiple and potentially incompatible relations between two terms. Three of such source vocabularies are further investigated in the following section. Finally, analysis at the term level also shows that the source vocabularies are responsible for the presence of multiply-related concepts in the UMLS only in a quarter of cases.
Our study has several limitations. First, it concentrates on precise situations within the UMLS. Because these correspond to complex relation combinations, they represent a small percentage of the entire set of UMLS relations. Second, our study is limited to multiply-related concepts when they are distinct. It would be interesting to investigate multiple relationships existing within a unique concept. Finally, our analysis at the term level may have underestimated the responsibility of source vocabularies for the presence of multiple relations between the investigated concepts. In particular, MDR uses distinct terms (and codes) to represent a unique concept,44 which may result in misinterpretations. As an illustration, Agranulocytosis (C0001824) and Neutropenia (C0027947) are related through PAR and SIB in the UMLS because the MDR term Agranulocytosis (MDR code: 10001507) is associated with Neutropenias (10029355) and Neutropenia (10029354) through PAR and SIB, respectively. Here, the actual reason why multiple relations appear between the corresponding concepts is because MDR does not cluster synonymous terms into a unique code.
Analysis of source vocabularies that give rise to non-homogenous combinations at the term level
As seen in the ‘Reason for the presence of multiple relations between concepts’ section, seven source terminologies principally relate terms with non-homogeneous combinations, which may be problematic. We study three of them in more detail by analyzing the combinations of source relationships.
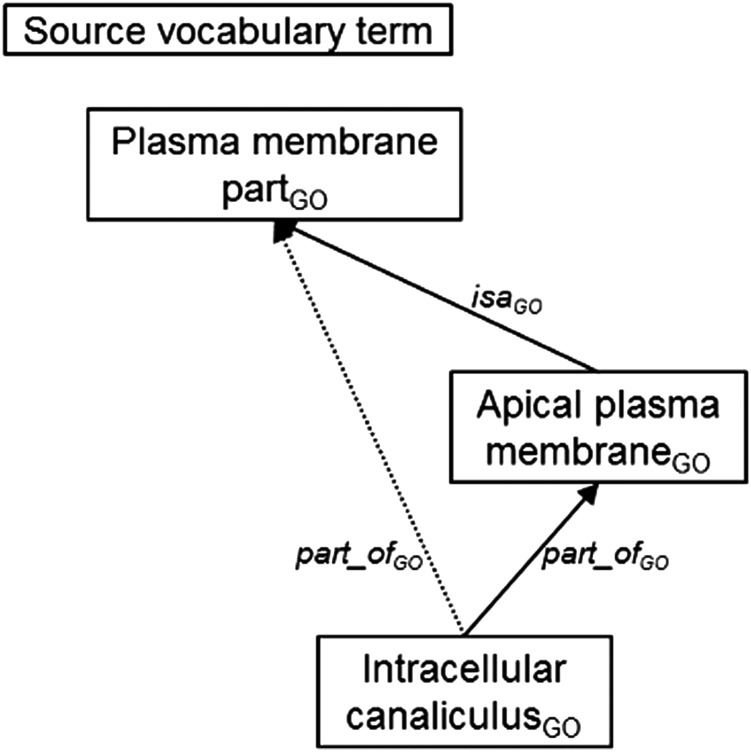
In GO,45 the most frequent combination is RB SIB, which is principally associated with the has_part none combination of source relationships. ‘None’ appears when a given relationship is not specified by a source vocabulary. Here, this is due to the presence of the relationship SIB, which is systematically added by the UMLS when two concepts have a common parent in a given source terminology. The relationship has_part is assigned to RB within the UMLS. When investigating examples of such combinations, we have observed redundant relations in GO, which then result in multiple relations within the UMLS. As an illustration from GO (figure 4), Intracellular canaliculus is represented as part_of Apical plasma membrane. In addition, Intracellular canaliculus is also defined as part_of Plasma membrane part, to which Apical plasma membrane is related through isa. The UMLS records a SIB relationship between Intracellular canaliculus (C0230646) and Apical plasma membrane (C1167182) because they are both associated with Plasma membrane part (C1820065) (although in the first case it is through a partitive relationship, whereas it is a subsumption one in the second case). As indicated in GO,46 if A part_of B and B isa C, then A part_of C. Thus, the relation part_of existing between Intracellular canaliculus and Plasma membrane part should be removed from GO because it can be inferred. Without this redundant relation, no SIB relationship would be recorded in the UMLS and only a single relation would exist between the corresponding concepts.
Figure 4.
A redundant partitive relation (dotted line) described in GO.
In NCI,47 the two major combinations of source relationships are inverse_isa parent_is_cdrh and inverse_isa is_biochemical_function_of_gene_product, both corresponding to the combination PAR RO. In any case, inverse_isa is assigned to PAR, while the second source relationship is assigned to RO. The relationship parent_is_cdrh is defined as a property ‘created to allow the source CDRH to assign a parent to each concept with the intent of creating a hierarchy that includes only terms in which they are the contributing source.’ According to this definition, the UMLS apparently misinterprets these relationships, which should preferably be assigned to PAR. Conversely, NCI surprisingly (and probably wrongly) combines inverse_isa with is_biochemical_function_of_gene_product for more than 1000 pairs of terms. An example is Interleukin-13 (C0214743) and Interleukins (C0021764), which are both gene products and should not be related through is_biochemical_function_of_gene_product.
In MEDLINEPLUS,48 the only combination of source relationships is related_to none, corresponding to the combination RO SIB. This situation occurs because MEDLINEPLUS is not a real terminology. Indeed, it provides information about high level subject categories for consumer health information, which group together medical topics associated only through related_to relationships, although more precise relationships would sometimes be more appropriate. As an illustration, Spina Bifida (C0080178) and Neural Tube Defects (C0027794) are associated through related_to in MEDLINEPLUS (RO) and, because they belong to the same medical topic group Genetics/Birth Defects (C1456603), a SIB relationship is also recorded within the UMLS between these two concepts. Actually, Spina Bifida should be described as isa Neural Tube Defects (its MEDLINEPLUS definition begins with ‘It is a type of neural tube defect’).
Preventing contradictory relationships
It should be noted that the presence of multiple relationships within the UMLS is not problematic per se, especially because its objective is to preserve all the relations asserted in the source vocabularies. However, this situation becomes problematic when the relations between two concepts are contradictory. As shown in the sections ‘Reason for the presence of multiple relations between concepts’ and ‘Detailed analysis of contradictory combinations,’ incompatible relationships are predominantly generated during the UMLS integration process and we mentioned multiple reasons why this happens. To avoid such situations, a simple solution could be to create a new concept if symmetric hierarchical relationships co-occur. This could solve cases like that presented in figure 3B: with an additional concept containing Amebiasis, unspecified, there would no longer be a granularity difference between relationships. Nevertheless, this solution may cause a dramatic increase of the number of concepts within the UMLS and may not solve all the problematic situations generated during the integration process. Thus, a clear identification of the problem, similar to that proposed here, is required so that users are aware of such situations and consider them cautiously.
In some cases, however, some source vocabularies define contradictory combinations between terms. For example, SNOMEDCT relates Pleural membrane structure (C0032225) and Entire pleura (C1279036) through inverse_isa and part_of (recorded as PAR and RN within the UMLS, respectively), although this appears to be contradictory. We suggest that such relations should be removed from source terminologies.
Conclusion
Our study is different from previous proposals in several ways: (1) we focus on multiply-related concepts; (2) we propose automatic and manual analyses of synonymous, hierarchical and associative relations; (3) we investigate the relationships’ compatibility; (4) and we study these situations at the UMLS scale and at the scale of source vocabularies (concept and term levels). The source terminologies are actually responsible for the presence of multiply-related concepts in the UMLS in a quarter of cases. The manual analysis was useful for explaining the conceptualization difference in relations between terms across source vocabularies. Finally, the exploitation of source relationships was helpful for understanding why some source terminologies describe multiple relations between a given pair of terms.
Acknowledgments
The authors would like to thank reviewers for their helpful suggestions.
Footnotes
Correction notice: This article has been corrected since it was published Online First. Figure 4 has been corrected.
Contributors: FM suggested and performed data analyses, performed detailed analysis of source vocabularies which cause non-homogenous combinations at the term level, and helped write the paper. NG had the original idea for this work, manually analyzed 288 pairs of multiply-related concepts, and helped write the paper. Both authors approved the submitted version of the manuscript.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Cabré M, Estopà R, Vivaldi J. Automatic term detection: a review of current systems. Recent advances in computational terminology. John Benjamins Publishing Company, 2001:53–88 [Google Scholar]
- 2.Qi G, Haase P, Huang Z, et al. A kernel revision operator for terminologies—algorithms and evaluation. Proceedings of the International Conference on the Semantic Web 2008:419–34 [Google Scholar]
- 3.Klein M, Fensel D. Ontology versioning on the semantic web. Proceedings of the Semantic Web Working Symposium 2001:75–91 [Google Scholar]
- 4.Maedche A, Motik B, Stojanovic L, et al. Managing multiple ontologies and ontology evolution in ontologging. Proceedings of the Symposium Intelligent Information Processing 2002:51–63 [Google Scholar]
- 5.D'aquin M, Euzenat J, Le Duc C, et al. Sharing and reusing aligned ontologies with cupboard. Proceedings of Knowledge Capture 2009:179–80 [Google Scholar]
- 6.Lindberg D, Humphreys B, McCray A. The Unified Medical Language System. Methods Inf Med 1993;32:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32:D267–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fellbaum C. A semantic network of English: the mother of all WordNets. EuroWordNet: a multilingual database with lexical semantic network. Computers and the Humanities 1998;32:209–20 [Google Scholar]
- 9.Fischer DH. Formal redundancy and consistency checking rules for the lexical database WordNet 1.5. Proceedings of the ACL Workshop on Automatic Information Extraction and Building of Lexical Semantic Resources for NLP Applications 1997:22–31 [Google Scholar]
- 10.Devitt A, Vogel C. The topology of WordNet: some metrics. Proceedings of the Global WordNet Conference 2004:106–11 [Google Scholar]
- 11.Smrz P. Quality control for WordNet development. Proceedings of the Global WordNet Conference 2004:206–12 [Google Scholar]
- 12.Liu Y, Yu J, Wen Z, et al. Two kinds of hypernymy faults in WordNet: the cases of ring and isolator. Proceedings of the Global WordNet Conference 2004:347–51 [Google Scholar]
- 13.Zhu X, Fan J, Baorto D, et al. A review of auditing methods applied to the content of controlled biomedical terminologies. J Biomed Inform 2009;42:413–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimino J. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998;37:394–403 [PMC free article] [PubMed] [Google Scholar]
- 15.Cimino J. Auditing the Unified Medical Language System with semantic methods. J Am Med Inform Assoc 1998;5:41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu H, Perl Y, Elhanan G, et al. Auditing concept categorizations in the UMLS. Artif Intell Med 2004;31:29–44 [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Johnson S, Friedman C. Automatic resolution of ambiguous terms based on machine learning and conceptual relations in the UMLS. J Am Med Inform Assoc 2002;9:621–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphreys B, McCray A, Cheh M. Evaluating the coverage of controlled health data terminologies: report on the results of the NLM/AHCPR large scale vocabulary test. J Am Med Inform Assoc 1997;4:484–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cimino J. Representation of clinical laboratory terminology in the Unified Medical Language System. Proceedings of Computer Application in Medical Care 1991:199–203 [PMC free article] [PubMed] [Google Scholar]
- 20.Moss J, Coenen A, Mills M. Evaluation of the draft international standard for a reference terminology model for nursing actions. J Biomed Inform 2003;36:271–8 [DOI] [PubMed] [Google Scholar]
- 21.Penz J, Brown S, Carter J, et al. Evaluation of SNOMED coverage of veterans health administration terms. Stud Health Technol Inform 2004;107(Pt 1):540–4 [PubMed] [Google Scholar]
- 22.Huang K, Geller J, Halper M, et al. Using WordNet synonym substitution to enhance UMLS source integration. Artif Intell Med 2008;46:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cimino J, Johnson S, Hripcsak G, et al. Managing vocabulary for a centralized clinical system. Medinfo 1995;8(Pt 1):117–20 [PubMed] [Google Scholar]
- 24.Chute C, Cohn S, Campbell K, et al. The content coverage of clinical classifications for the computer-based patient record institute's work group on codes & structures. J Am Med Inform Assoc 1996;3:224–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald F, Chute C, Ogren P, et al. A large-scale evaluation of terminology integration characteristics. Proc AMIA Symp 1999:864–7 [PMC free article] [PubMed] [Google Scholar]
- 26.Bodenreider O, Mitchell J, McCray A. Evaluation of the UMLS as a terminology and knowledge resource for biomedical informatics. Proc AMIA Symp 2002:61–5 [PMC free article] [PubMed] [Google Scholar]
- 27.Bodenreider O, Burgun A, Rindflesch T. Assessing the consistency of a biomedical terminology through lexical knowledge. Int J Med Inform 2002;67:85–95 [DOI] [PubMed] [Google Scholar]
- 28.Cimino J, Hripcsak G, Johnson S, et al. Prototyping a vocabulary management system in an object-oriented environment. Proceedings of IMIA WG Software Engineering in Medical Informatics 1990:429–39 [Google Scholar]
- 29.Cimino J, Clayton P. Coping with changing controlled vocabularies. Proc Annu Symp Comput Appl Med Care 1994:135–9 [PMC free article] [PubMed] [Google Scholar]
- 30.Bodenreider O, Burgun A, Botti G, et al. Evaluation of the unified medical language system as a medical knowledge source. J Am Med Inform Assoc 1998;5:76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geller J, Gu H, Perl Y, et al. Semantic refinement and error correction in large terminological knowledge bases. Data Knowl Eng 2003;45:1–32 [Google Scholar]
- 32.Bodenreider O. Strength in numbers: exploring redundancy in hierarchical relations across biomedical terminologies. Proc AMIA Symp 2003:101–5 [PMC free article] [PubMed] [Google Scholar]
- 33.Gu H. Evaluation of a UMLS auditing process of semantic type assignments. Proc AMIA Symp 2007:294–8 [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz E, Barrett J, Price C. Semantic quality through semantic definition: refining the Read Codes through internal consistency. Proc AMIA Symp 1997:615–9 [PMC free article] [PubMed] [Google Scholar]
- 35.Burgun A, Bodenreider O. Methods for exploring the semantics of the relationships between co-occurring UMLS concepts. Stud Health Technol Inform 2001;84:171–5 [PMC free article] [PubMed] [Google Scholar]
- 36.Wang A, Sable J, Spackman K. The SNOMED clinical terms development process: refinement and analysis of content. Proc AMIA Symp 2002:845–9 [PMC free article] [PubMed] [Google Scholar]
- 37.Cornet R, Abu-Hanna A. Description logic-based methods for auditing frame-based medical terminological systems. Artif Intell Med 2005;34:201–17 [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Halper M, Perl Y, et al. Relationship structures and semantic type assignments of the UMLS enriched semantic network. J Am Med Inform Assoc 2005;12:657–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min H, Perl Y, Chen Y, et al. Auditing as part of the terminology design life cycle. J Am Med Inform Assoc 2006;13:676–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Halper M, Min H, et al. Structural methodologies for auditing SNOMED. J Biomed Inform 2007;40:561–71 [DOI] [PubMed] [Google Scholar]
- 41.Cornet R, Abu-Hanna A. Auditing description-logic-based medical terminological systems by detecting equivalent concept definitions. Int J Med Inform 2008;77:336–45 [DOI] [PubMed] [Google Scholar]
- 42.Gu H, Elhanan G, Halper M, et al. Questionable relationship triplet in the UMLS. Proceedings of the IEEE-EMBS International Conference on Biomedical and Health Informatics 2012:713–6 [Google Scholar]
- 43.Grabar N, Dupuch M, Mougin F. Dommages collatéraux de la fusion de terminologies. Proceedings of the Ninth International Conference on Terminology and Artificial Intelligence 2011:10–16 [Google Scholar]
- 44.Merrill G. The MedDRA paradox. Proc AMIA Symp 2008:470–4 [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. http://www.geneontology.org/GO.ontology.relations.shtml.
- 47.Hartel FW, De Coronado S, Dionne R, et al. Modeling a description logic vocabulary for cancer research. J Biomed Inform 2005;38:114–29 [DOI] [PubMed] [Google Scholar]
- 48.Miller N, Lacroix EM, Backus JE. MEDLINEplus: building and maintaining the National Library of Medicine's consumer health Web service. Bull Med Libr Assoc 2000;88:11–17 [PMC free article] [PubMed] [Google Scholar]