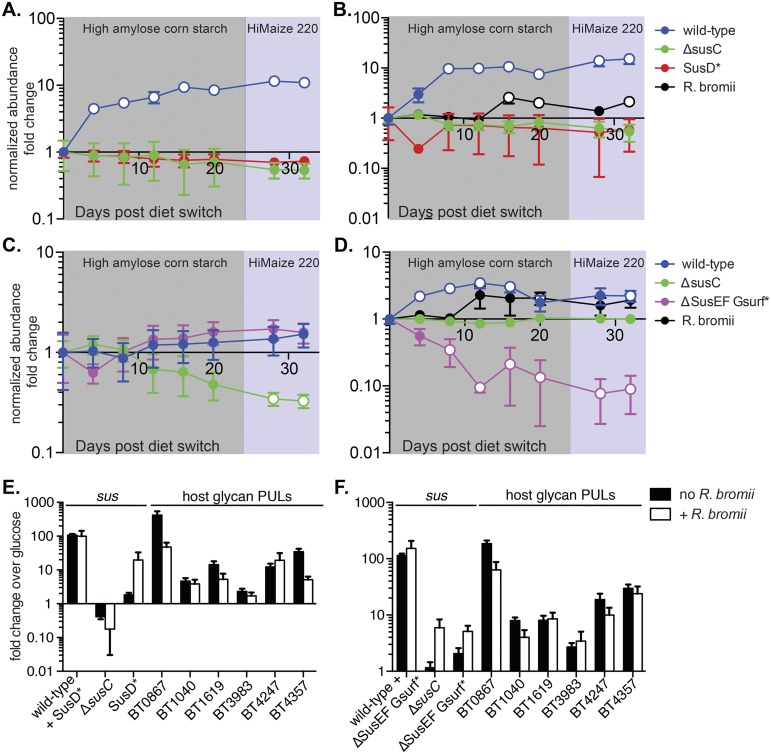
FIG 6 .
The Sus binding sites enhance wild-type B. thetaiotaomicron fitness in vivo on a starch-rich diet. Germfree mice fed a starch-free diet were randomly segregated into four treatment groups (n = 5 per group) and colonized with a mixture of wild-type and mutant B. thetaiotaomicron strains. All groups were colonized with wild-type, ΔsusC, and either SusD* (A, B) or ΔSusEF Gsurf* (C, D) strains. Half of the groups (B, D) were also colonized with the keystone starch-degrading species Ruminococcus bromii. Once colonization was established, mice were switched to a diet rich in high-amylose resistant cornstarch for 24 days (shaded in gray) and then switched to a diet rich in Hi-Maize 220 (shaded in purple) for the remainder of the experiment. The relative abundance of each strain was determined by quantifying unique genomic tags using qPCR and was normalized to its abundance on the day of the initial diet switch (day 0). Average results and standards errors across the five mice are shown. Open circles represent significant changes (P < 0.05) in normalized abundance versus the abundance on day 0, calculated using the two-tailed Student’s t test. (E and F) sus transcript levels from cecal contents collected at the end of the experiment were probed using qPCR, and fold changes relative to the levels of B. thetaiotaomicron grown in MM-glucose were calculated. (E) Transcripts from groups whose abundance results are shown in panels A and B. (F) Data from groups whose abundance results are shown in panels C and D. To probe for strain-specific sus expression, primers were designed such that sus transcripts would be amplified from only a subset of the B. thetaiotaomicron strains present (noted on the x axis). sus expression levels were normalized to the relative abundance of the strain from which they were amplified. Transcript levels of PULs targeting host mucosal glycans were probed as well.