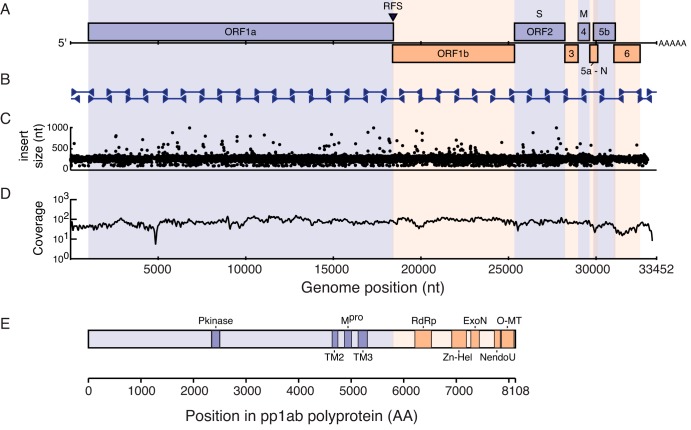
FIG 4 .
Ball python nidovirus genome and replicase polyprotein organization. (A) A cartoon showing the genome organization of ball python nidovirus. Major open reading frames are indicated. RFS, position of putative ribosomal frameshift “slippery” sequence. (B) Position of Sanger sequencing PCR and RACE amplicons used to validate the NGS-based assembly (C) Concordance plot of read pairs mapping to assembly. Read pairs from snake no. 1 data set were mapped to the genome assembly using the Bowtie2 aligner. The inferred size of each library molecule is plotted. (D) Read coverage. Reads were aligned as in panel C, and the average number of bases supporting each position in 50-nt sliding windows of the alignment is indicated. The dip at approximately nt 5000 corresponds to a repeat-containing region with decreased mappability. (E) Conserved domains present in the replicase polyproteins (pp1ab). Domains were identified by searching the PFam database using the HMMER3 tool as described in the text and Table 2. Scale bars are indicated.