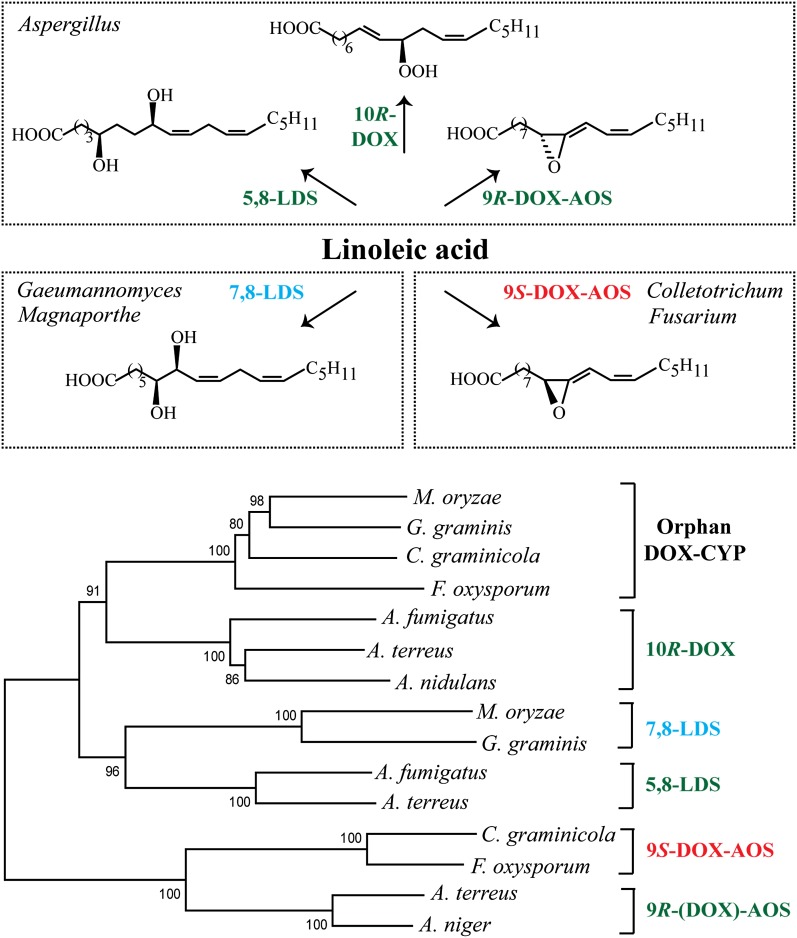
Fig. 1.
Overview of oxylipin biosynthesis by the genera Aspergillus, Gaeumannomyces, Magnaporthe, Colletotrichum, and Fusarium and phylogenetic tree of DOX-CYP subfamilies. A: Linoleic acid is oxidized by 5,8-LDS, 10R-DOX, and 9R-DOX-AOS of A. terreus and other aspergilli (top). Oxylipins formed from 18:2n-6 by 7,8-LDS of Gaeumannomyces and Magnaporthe (e.g., G. graminis, M. oryzae) and by 9S-DOX-AOS of F. oxysporum and C. graminicola (bottom). B: Phylogenetic tree of MGG_10859 (M. oryzae) and three homologous sequences (EJT82559, EFQ36272, and FOXB_03425, respectively) with >60% amino acid identity covering >90% of the sequence. The next sequences are from top to bottom: 10R-DOX (ACL14177, ATEG_04755, and AAT36614), 7,8-LDS (MGG_13239, Q9UUS2), 5,8-LDS (ACO55067, ATEG_03992), 9S-DOX-AOS (EFQ27323, FOXB_01332), and 9R-(DOX)-AOS [ATEG_02036, EHA25900 (Hoffman and Oliw, unpublished observation)]; the parentheses in 9R-(DOX)-AOS emphasize that recombinant ATEG_02036, in contrast to recombinant EHA25900, lacks the 9R-DOX activities.