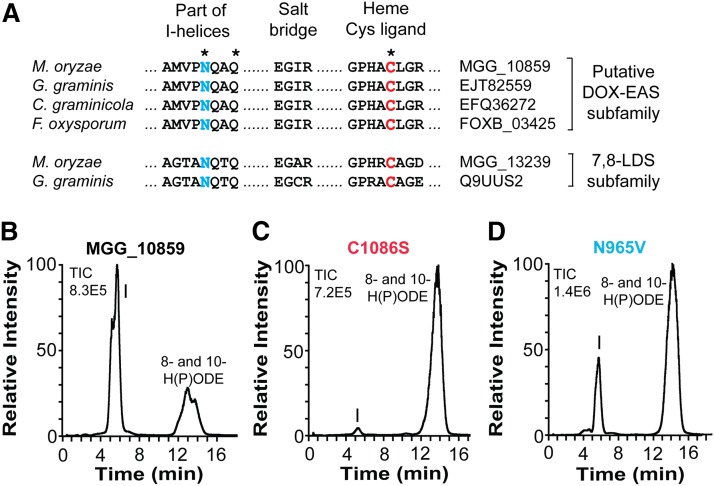
Fig. 4.
Comparison of partial sequences of four members of the 10R-DOX-EAS subfamily with 7,8-LDS of M. oryzae and G. graminis and effects of site-directed mutagenesis of Cys and Asn residues. A: The alignment covers the conserved AsnGlnXaaGln sequences, which are believed to be important for substrate positioning, the GluXaaXaaArg (“EXXR”) sequences, functioning as a salt bridge, and the motif with the heme thiolate ligand. The latter two sequences are typical of P450s. Asterisks indicate the three amino acid residues of MGG_10589, which were subject to site-directed mutagenesis. The sequence identity within the putative 10R-DOX-EAS subfamily is 60% or higher. B: RP-HPLC analysis of products formed from 18:2n-6 by 10R-DOX-EAS (MGG_10859). C: RP-HPLC/MS analysis of products formed from 18:2n-6 by 10R-DOX-EAS•C1086S. D: RP-HPLC/MS analysis of products formed from 18:2n-6 by 10R-DOX-EAS•N965V. Peak I designates 12S(13R)-epoxy-10(R)-hydroxy-8(E)-octadecenoic acid, which was identified by MS2 analysis in all three chromatograms.