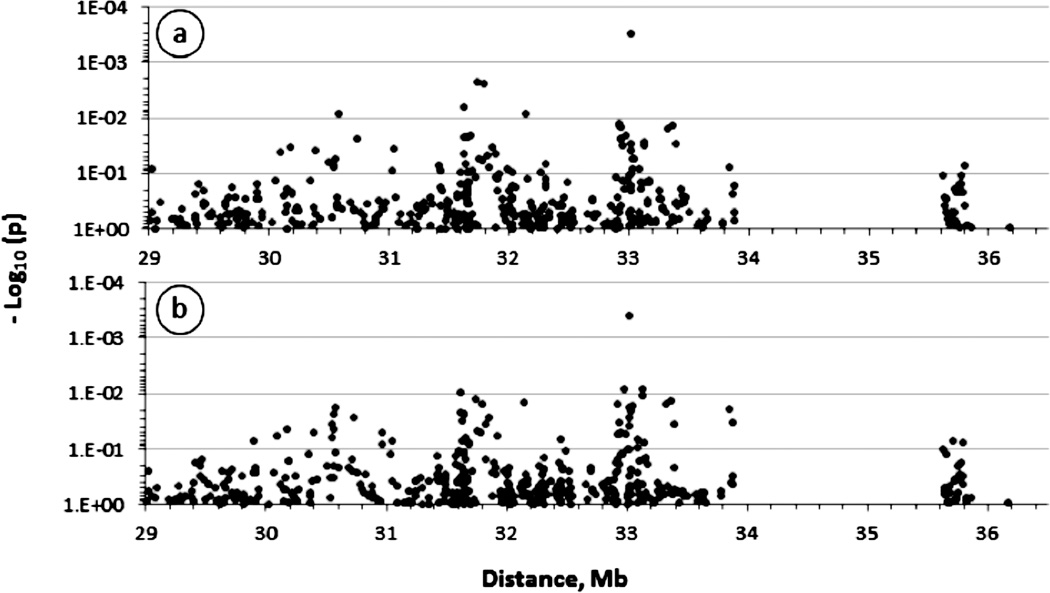
Figure 2.
Association of major histocompatibility complex (MHC) polymorphisms with HIV-related Kaposi’s sarcoma (HIV-KS) in men. The figure shows the strength of the association expressed as minus logarithm decimal of the p-value (−Log 10(p)) obtained from univariate analyses of 467 single nucleotide polymorphisms (SNP) spanning about 5 megabases across central and extended MHC. The association with the KS outcome was evaluated in conditional logistic models assuming additive variance and controlling for the effects of age at baseline, degree of immunosuppression (calculated as the area under the curve of CD4+ T-cell count below 300) and the rate of CD4+ T-cell count change at six month-intervals (slope of CD4+ T-cell count from the time of dual seroconversion to HIV-1 and HHV8). In panel B, the logistic models were further controlled for the effects of two HLA-B alleles (B*2705 and B*1401) significantly (p<5%) associated with KS. The large centromeric gap corresponds to a chromosome 6p21genomic interval where no genes of potential relevance to KS were found at the time this study was initiated.