FIGURE 5.
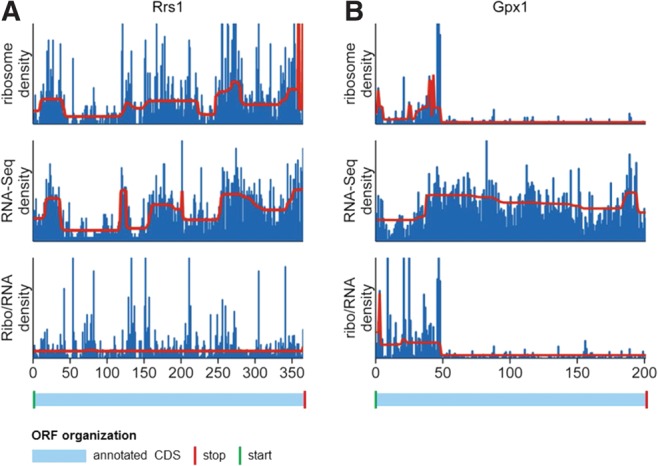
Elimination of artifacts from ribosome profiling. RNA read densities are shown in blue, and the segments of equal density estimated by the change point (CP) algorithm are shown in red. After normalization by RNA-Seq, artifacts, such as effects of multiple alignments and assignments, are removed from ribosome profiles. (A) For Rrs1, normalization removed all artifacts from the ribosome profile; consequently, the CP analysis of the normalized profile resulted in no CPs found. (B) For Gpx1, a single CP remained after normalization, which we later showed to be a true translation regulation event. The annotated CDSs are shown below the density plots in light blue. Green vertical lines indicate annotated start codons, red lines indicate annotated stop codons, and black lines indicate exon junctions.
