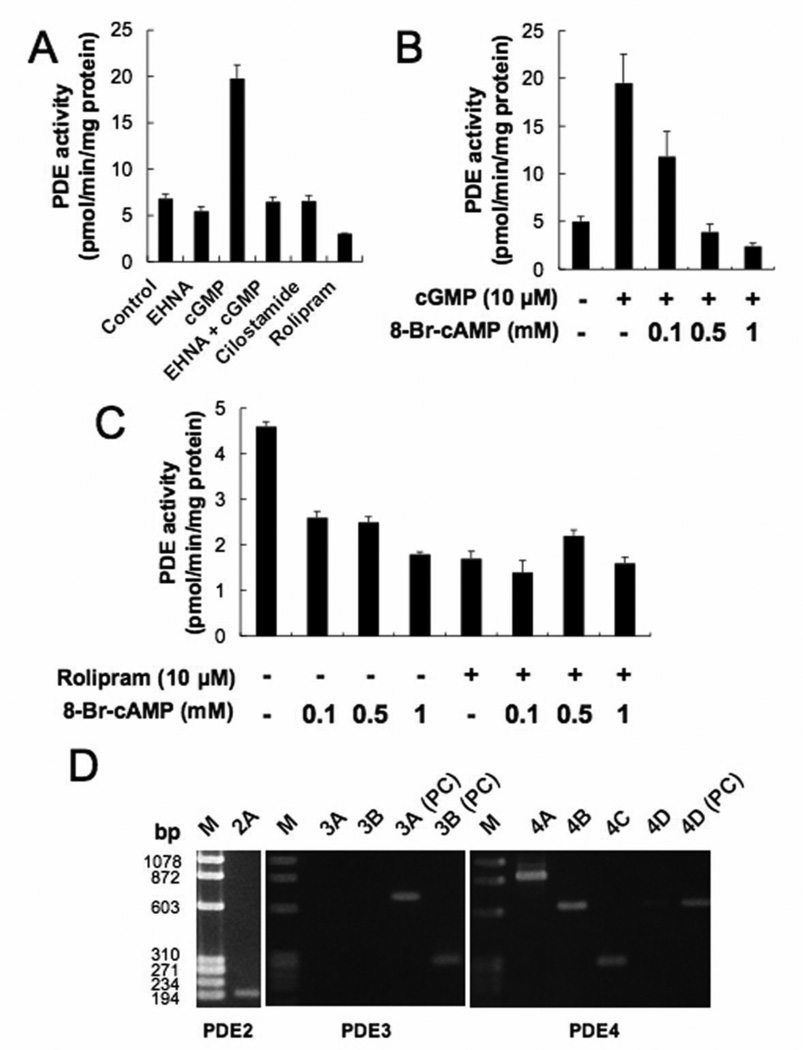
Fig. 2.
Expression of PDEs and effects of 8-Br-cAMP on PDE activity in PMP cells. Data in graphs are means of three independent experiments, each performed in triplicate. (A) PDE activities were analyzed by cAMP PDE activity assay with or without each specific PDE inhibitor. The error bars represent means ± SD (n = 3). The concentrations of each reagents were: EHNA, 20 µM; cGMP, 10 µM; cilostamide, 0.5 µM; rolipram, 10 µM. (B) Effect of 8-Br-cAMP on cGMP-stimulated PDE activity in PMP cells. cGMP (10 µM) and 8-Br-cAMP (0.1 to 1 mM) were used. The error bars represent means ± SD, n = 3. (C) Effect of 8-Br-cAMP with or without rolipram on PDE activity in PMP cells. Rolipram (10 µM) and 8-Br-cAMP (0.1 to 1 mM) were used. (D) Expression of PDE mRNAs in PMP cells. RT-PCR analysis for PDE2, PDE3, and PDE4 mRNAs were performed. HMG cells derived from human gingival malignant melanoma were used as the positive control (PC) for PDE3A, 3B, and 4D mRNAs. Experiments were repeated three times, and similar results were obtained. 2A = PDE2A; 3A = PDE3A; 3B = PDE3B; 4A = PDE4A; 4B = PDE4B; 4C = PDE4C; 4D = PDE4D; M = molecular markers.