Abstract
Mitochondrial dysfunction is associated with neuronal loss in Huntington’s disease (HD), a neurodegenerative disease caused by an abnormal polyglutamine expansion in huntingtin (Htt). However, the mechanisms linking mutant Htt and mitochondrial dysfunction in HD remain unknown. We identify an interaction between mutant Htt and the TIM23 mitochondrial protein import complex. Remarkably, recombinant mutant Htt directly inhibited mitochondrial protein import in vitro. Furthermore, mitochondria from brain synaptosomes of presymptomatic HD model mice and from mutant Htt-expressing primary neurons exhibited a protein import defect, suggesting that deficient protein import is an early event in HD. The mutant Htt-induced mitochondrial import defect and subsequent neuronal death were attenuated by overexpression of TIM23 complex subunits, demonstrating that deficient mitochondrial protein import causes mutant Htt-induced neuronal death. Collectively, these findings provide evidence for a direct link between mutant Htt, mitochondrial dysfunction and neuronal pathology, with implications for mitochondrial protein import-based therapies in HD.
HD is a fatal autosomal dominant neurodegenerative disorder characterized by progressive neurological dysfunction and selective neuronal loss in the striatum and cortex1. Although the genetic cause of HD, an abnormal expansion of CAG repeats encoding polyglutamine (polyQ) in exon 1 of the HTT gene, has been identified2, the pathogenic mechanisms remain unclear, and no current therapy ameliorates the neurodegenerative process.
Mitochondrial dysfunction has been highlighted as a critical driver of HD pathophysiology3–5. Mitochondria are important in diverse cellular functions, including bioenergetics, calcium homeostasis and apoptotic signaling. Several proteolytically cleaved N-terminal fragments of mutant Htt proteins have been identified in cells and appear to be more cytotoxic and prone to aggregation than full-length mutant Htt6–8. Ultrastructural and biochemical evidence indicates that N-terminal fragments of mutant Htt associate with mitochondria in cellular and animal models of HD9–11, suggesting that mutant Htt directly affects mitochondrial function. However, the mechanism directly linking mutant Htt and mitochondrial dysfunction remains unknown.
Mitochondria contain approximately 1,500 different proteins, 99% of which are encoded by the nuclear genome12. Therefore, the import, sorting and assembly of nuclearly encoded mitochondrial proteins are essential for normal mitochondrial function. Only 13 proteins of the respiratory chain are encoded by the mitochondrial genome and synthesized in mitochondria. Nuclearly encoded mitochondrial proteins are synthesized in cytosolic ribosomes as precursor proteins and imported into mitochondria by evolutionarily conserved multi-subunit mitochondrial membrane translocases: translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM)12,13. Whereas the TOM complex serves as the entry gate for almost all nuclearly encoded proteins, two distinct TIM complexes, the TIM23 and TIM22 complexes, act in the inner membrane. The TIM23 complex imports all matrix proteins and a subset of inner membrane and intermembrane space proteins, which harbor N-terminal cleavable presequences. The TIM22 complex, a carrier translocase, imports hydrophobic inner membrane proteins through internal targeting signals. Thus, nuclearly encoded mitochondrial proteins use specific import systems for precise mitochondrial localization. Blockade of import pathways is believed to lead to mitochondrial dysfunction14.
Here we demonstrate that mutant Htt localizes to brain mitochondria in human HD. Mutant Htt specifically associates with the TIM23 complex and directly inhibits protein import in isolated brain mitochondria. In HD mice, we observed a defect in protein import early in the disease in forebrain synaptosomal mitochondria, but not liver mitochondria. In addition, primary neurons expressing mutant Htt exhibited impaired mitochondrial protein import. Inhibition of protein import was sufficient to trigger neuronal death, and augmentation of protein import rescued mutant Htt–expressing neurons from cell death. Thus, deficient mitochondrial protein import is an early, tissue-specific, mutant Htt–induced pathogenic defect leading to neuronal death.
RESULTS
Mutant Htt binds to the mitochondrial import machinery
Mutant Htt associates with mitochondria in the brain of various HD transgenic mice9,10,15,16. To determine whether mutant Htt protein localizes to mitochondria in human brains affected by HD, we examined the caudate nucleus, the area most severely affected, from patients with grade 2 HD. Brain sections were subjected to immunohistochemistry with antibodies recognizing mitochondrial resident proteins, including a mitochondrial inner membrane translocase subunit, Tim23 and dynamin-related protein 1 (DRP1), and aggregated mutant Htt. Confocal immunofluorescence microscopy revealed localization of aggregated mutant Htt to mitochondria (Fig. 1a). Additionally, confocal microscopy identified partial colocalization of mutant Htt with mitochondrially targeted GFP (mtGFP) in mutant Htt knock-in mouse striatal cells (ST-HdhQ111/Q111) (Fig. 1b). These results suggest that mutant Htt may affect mitochondrial function by interacting with specific mitochondrial proteins.
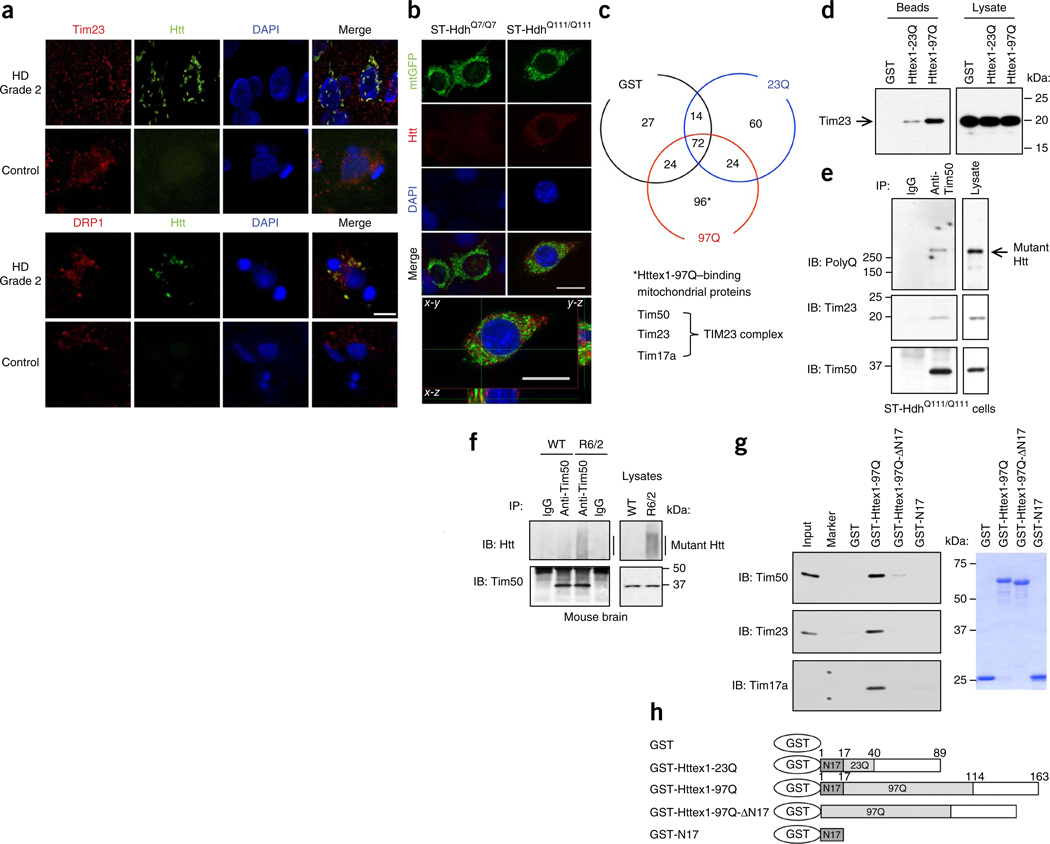
Figure 1.
Mutant Htt interacts with the TIM23 complex. (a) Caudate nucleus sections from human HD grade 2 and control brains, subjected to immunohistochemistry for indicated proteins. Mutant Htt aggregates detected by anti-Htt (EM48) antibody colocalize with mitochondrial proteins Tim23 and DRP1 in human HD caudate nucleus in deconvolved confocal images. (b) ST-Hdh cells transfected with mtGFP expression plasmid, subjected to immunofluorescence with anti-polyQ antibody (1C2) to label mutant Htt (red). Mutant Htt in ST-HdhQ111/Q111 cells partially colocalizes (yellow) with mitochondria in deconvolved confocal images. Scale bars (a,b), 10 µm. DAPI, 4′,6-diamidino-2-phenylindole. (c) Mouse forebrain mitochondria incubated with GST alone or GST-Httex1 proteins were subjected to GST pull-down assays. Bound proteins were identified by mass spectrometric analysis. Venn diagram represents the number of identified proteins. TIM23 complex components were identified among the Httex1-97Q–binding proteins. See also Supplementary Table 1. (d) Interaction between Tim23 and Httex1-97Q was verified by GST pull-down assays as in c, followed by immunoblotting with anti-Tim23 antibody. (e,f) Endogenous interaction between mutant Htt and Tim50 was found in ST-HdhQ111/Q111 cells (e) and 5-week-old R6/2 mouse forebrain (f) by coimmunoprecipitation (IP) with anti-Tim50 antibody followed by immunoblotting (IB). Normal IgG is a negative control. (g) Mitochondria isolated from ST14A cells were incubated with equimolar concentration of the indicated recombinant GST-fusion proteins and were subjected to GST pull-down assays. Bound proteins were analyzed by immunoblotting with indicated antibodies. Marker lane shows 20- and 15-kDa molecular mass standards. Right, Coomassie blue staining of GST fusion proteins used in assays. (h) A schematic representation of GST-Htt proteins used in c,d,g. The experiments (a,b,d–g) were successfully repeated three times. Full-length blots/gels (d–g) are presented in Supplementary Figure 9.
To identify mitochondrial proteins that form a complex with mutant Htt, we used a biochemical approach and performed a pull-down experiment using a recombinant mutant Htt exon 1 (Httex1) N-terminal fragment fused to glutathione S-transferase (GST). We incubated purified mouse forebrain mitochondria with GST alone or GST fusion proteins containing Httex1 with a normal (GST-Httex1-23Q) or pathological (GST-Httex1-97Q) polyQ repeat, and subjected bound proteins to mass spectrometry (Fig. 1c). We identified 96 proteins that bound only to Httex1-97Q, but not to GST alone or GST-Httex1-23Q (Fig. 1c and Supplementary Table 1). Among the Httex1-97Q–specific binding proteins, we found Tim50, Tim23 and Tim17a, all of which are subunits of the TIM23 complex in the inner membrane (Fig. 1c). Notably, although we detected several subunits of the TIM23 complex, we detected no subunits of the TIM22 carrier translocase complex, suggesting a specific interaction of mutant Htt with the TIM23 import pathway. All other proteins identified in the analysis are presented in Supplementary Table 1.
We verified interaction of mutant Htt with components of the TIM23 complex by GST pull-down assays using isolated forebrain mitochondria (Fig. 1d and data not shown). We found that the Tim23 subunit robustly bound to Httex1-97Q, with much weaker affinity for Httex1-23Q, but not to GST alone (Fig. 1d). An endogenous interaction between mutant Htt and TIM23 complex subunit Tim50 was detected in knock-in striatal cells expressing full-length mutant Htt and in R6/2 HD transgenic mouse brain, which expresses the mutant Httex1 N-terminal fragment (Fig. 1e,f). Together, these results suggest that the N-terminal portion of mutant Htt can associate with mitochondria through a specific interaction with the import complex.
It has been suggested that the first N-terminal 17 amino acids (N17) of Htt are key to its subcellular localization17. We therefore performed GST pull-down assays using purified GST-Httex1-97Q, GST-Httex1-97Q-ΔN17, which lacks the N17 sequence, and GST-N17 with mitochondria isolated from conditionally immortalized rat embryonic striatal ST14A cells and mouse brain. Both GST-Httex1-97Q-ΔN17 and GST-N17 showed little, if any, interaction with Tim17a, Tim23 and Tim50 (Fig. 1g,h and data not shown), suggesting that N17 is required but not sufficient for interaction with the TIM23 complex. These results suggest that both N17 and the polyQ domains of mutant Htt are critical for the interaction with the TIM23 import machinery. Moreover, compared to Tim23 and Tim50, Tim17a was most enriched by GST-Httex1-97Q pulldown, suggesting that Httex1-97Q may interact primarily with Tim17a within the TIM23 complex. Collectively, these findings suggest that mutant Htt associates with mitochondria through a specific and direct interaction with the TIM23 complex and raise the hypothesis that mutant Htt may interfere with the import machinery.
Mutant Htt inhibits brain mitochondrial protein import
Given the physical association of mutant Htt with a mitochondrial translocase (Fig. 1c–g), we reasoned that mutant Htt may act directly on mitochondria to inhibit protein import and took advantage of an established in vitro protein import assay with a radiolabeled precursor matrix protein, pre-ornithine transcarbamylase (pOTC), to examine import activity in normal mitochondria in the presence of recombinant GST-Htt fusion proteins in vitro. pOTC is translocated across the outer and inner mitochondrial membranes via the TOM and TIM23 complexes, respectively, and is destined for the matrix, where the N-terminal presequence in pOTC is cleaved by a mitochondrial processing peptidase, producing mature OTC (mOTC). We incubated isolated mitochondria with 35S-labeled pOTC and detected imported mOTC in the matrix by fluorography after SDS-PAGE. This assay reflects the import activity for many nuclearly encoded mitochondrial proteins, including presequence-containing matrix, inner membrane and intermembrane space proteins that also utilize the TOM and TIM23 complexes12.
We first incubated isolated mouse forebrain mitochondria with GST, GST-Httex1-23Q or GST-Httex1-97Q recombinant proteins (Fig. 2a) and then subjected them to the in vitro mitochondrial pOTC import assay (Fig. 2b,c). GST-Httex1-97Q proteins inhibited pOTC import in mitochondria as compared to GST alone or GST-Httex1-23Q (Fig. 2b,c). Incubation of wild-type (WT) mitochondria with 3 µM and 10 µM GST-Httex1-97Q reduced pOTC import by 50% and 73%, respectively, compared to that of control GST (Fig. 2b), indicating that the mutant Htt N-terminal fragment directly inhibits mitochondrial protein import in vitro. Incubation of mitochondria with GST-Httex1-23Q also led to a modest inhibitory effect on pOTC import as compared to incubation with GST (Fig. 2b,c), raising the possibility that normal Htt may regulate protein import in mitochondria. However, two lines of evidence suggest this is not the case in cells. First, we observed no difference in mitochondrial protein import activity in Htt-null embryonic stem cells as compared to WT embryonic stem cells or in Htt knockdown ST14A cells as compared to vector-transfected cells (Supplementary Fig. 1 and data not shown). Second, N-terminal WT Httex1-25Q does not localize to mitochondria in transfected neurons, whereas mutant Httex1-97Q does11, suggesting that, in cells, WT Htt is not poised to affect mitochondrial protein import. Together, our results suggest that mutant Htt directly inhibits mitochondrial protein import via interaction with the import machinery.
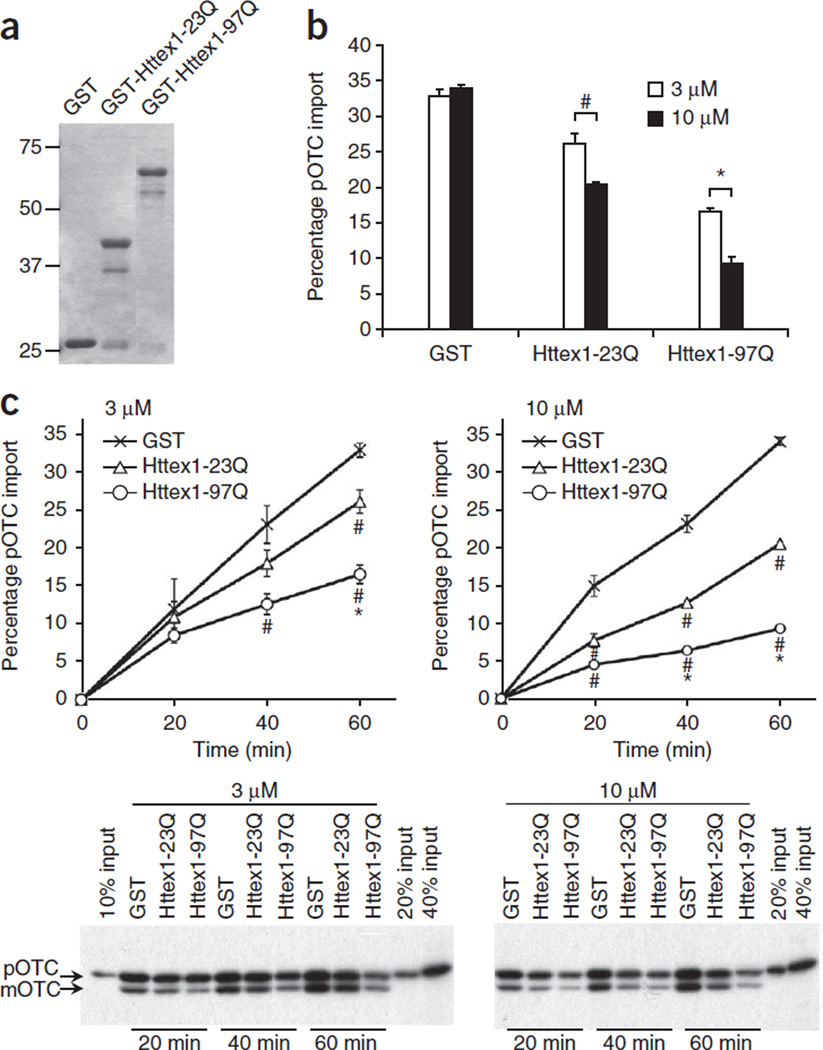
Figure 2.
Mutant Htt directly inhibits mitochondrial protein import. (a) Coomassie blue staining of GST fusion proteins used in b,c. (b) Forebrain mitochondria prepared from adult WT mice were preincubated with GST or GST-Httex1 proteins (3 or 10 µM) on ice for 1 h in 10 µl and then subjected to import assays by adding 40 µl import reaction buffer containing [35S]pOTC (60 min). Data are presented as mean + s.e.m. Inhibition of pOTC import by Httex1 proteins was dose dependent (#P = 0.001 and *P = 0.0001, F5,12 = 109.40). GST alone showed import activity similar to vehicle (phosphate-buffered saline), indicating no effect of GST on pOTC import (data not shown). (c) Kinetic analysis of pOTC import reaction after preincubation with indicated recombinant proteins as in b. Representative gel images used for quantification are shown. Import reaction times are indicated. Httex1 proteins decreased the import of pOTC into mitochondria in a polyQ length–dependent manner. # and * represent significant difference compared to GST and GST-Httex1-23Q at the given time point, respectively. 3 µM, 40 min: Httex1-97Q, #P = 0.008, F2,6 = 7.404. 3 µM, 60 min: Httex1-23Q, #P= 0.009; Httex1-97Q, #P< 0.0001, *P= 0.002, F2,6 = 43.28. 10 µM, 20 min: Httex1-23Q, #P= 0.002; Httex1-97Q, #P= 0.0002, F2,6 = 31.29. 10 µM, 40 min: Httex1-23Q, #P< 0.0001; Httex1-97Q, #P< 0.0001, *P= 0.0006, F2,6 = 153.73. 10 µM, 60 min: Httex1-23Q, #P< 0.0001; Httex1-97Q, #P< 0.0001, *P< 0.0001, F2,6 = 1,111.73. Data are presented as mean ± s.e.m. Percentage pOTC import (b,c) represents the percentage of mOTC radioactivity compared to input (total [35S]pOTC radioactivity added to each reaction). One-way ANOVA, Bonferroni t-test. n = 3 independent experiments (b,c). Full-length blots/gels (a,c) are presented in Supplementary Figure 9.
Mutant Htt impairs mitochondrial protein import in cells
Next we examined whether mutant Htt inhibits import in cells. The mutant Htt knock-in striatal cell line ST-HdhQ111/Q111, which expresses full-length mutant Htt protein, demonstrated decreased pOTC import as compared to control knock-in striatal line ST-HdhQ7/Q7, which expresses full-length WT Htt (Fig. 3a, left). We observed a similar deficit in import activity using mitochondria isolated from rat striatal ST14A cells that stably express the N-terminal 548-amino-acid fragment of mutant Htt with a 120Q repeat (N548mut) compared with that of cells expressing the N-terminal Htt fragment with a 15Q repeat (N548wt) (Fig. 3a, right). Finally, ST14A cells transiently transfected with plasmids encoding the N-terminal 170 amino acids of Htt with a pathological-length polyQ stretch (150Q) showed decreased import activity as compared with that of cells expressing Htt with a normal polyQ stretch (21Q) (Supplementary Fig. 2).
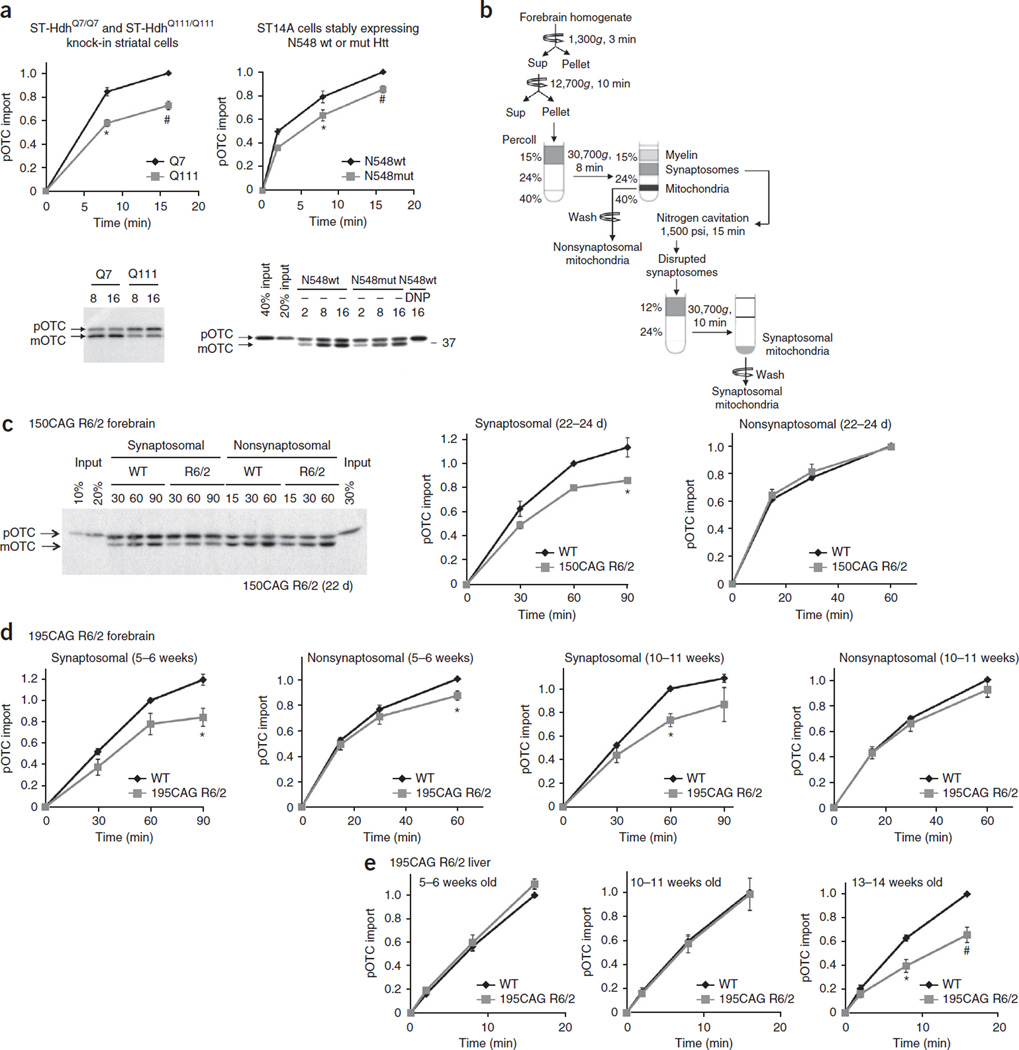
Figure 3.
Mitochondria isolated from mutant Htt-expressing striatal cells and mouse brain exhibit decreased protein import. (a) Mitochondria isolated from the indicated striatal cells were subjected to pOTC import assays. Addition of a mitochondrial uncoupler, 2,4-dinitrophenol (DNP), to mitochondria before starting the import reaction confirmed that dissipation of mitochondrial membrane potential blocks import. Mitochondria from ST-HdhQ111/Q111 and N548mut cells showed significantly decreased pOTC import compared to that of control cell lines. Times are indicated in minutes in a,c. ST-HdhQ111/Q111, 8 min: *P = 0.0004, t = 5.90, d.f. = 8, n = 5 independent experiments; ST-HdhQ111/Q111, 16 min: #P = 0.0022, U = 0, n = 6 independent experiments; N548mut, 8 min: *P = 0.045, t = 2.29, d.f. = 10, n = 6 independent experiments; N548mut, 16 min: #P = 0.0022, U = 0, n = 6 independent experiments. (b) Schematic of synaptosomal and nonsynaptosomal mitochondria isolation protocol from mouse forebrain. Sup, supernatant. (c) Synaptosomal mitochondria isolated from 22- to 24-d-old, presymptomatic 150CAG R6/2 mice showed significantly decreased pOTC import compared to that of WT (*P = 0.029, t = 3.34, d.f. = 4, n = 3 independent experiments; 3 or 4 WT or R6/2 brains were pooled in each experiment). (d) Synaptosomal mitochondria isolated from 195CAG R6/2 showed significantly decreased pOTC import compared to that of WT (5–6 weeks, *P = 0.012, t = 3.59, d.f. = 6, n = 4 independent experiments; 10–11 weeks, *P = 0.029, U = 0, n = 4 independent experiments). Modest reduction of pOTC import was also found in 195CAG R6/2 nonsynaptosomal mitochondria at 5–6 weeks of age compared to that of control WT (*P = 0.0006, U = 0, n = 7 independent experiments), but not at 10–11 weeks of age (n = 9 independent experiments). (e) 195CAG R6/2 liver mitochondria showed significantly impaired pOTC import only in late-disease stage (13–14 weeks) (*P = 0.0044, t = 3.92, d.f. = 8; #P = 0.0079, U = 0, n = 5 independent experiments). (a,e) The data are scaled to pOTC import (equal to mOTC) in control mitochondria after the maximum reaction time (set equal to 1). (c,d) The data are scaled to mOTC in WT mitochondria at 60 min reaction time (set equal to 1). (a,c) Representative gel images of at least 3 independent experiments are shown. (a,c–e) Mann-Whitney U and unpaired t-tests (two-tailed). Data are presented as mean ± s.e.m. Full-length blots/gels (a,c) are presented in Supplementary Figure 9.
In addition to pOTC import assays, we measured mitochondrial protein import in living cells expressing mtGFP, which contains a presequence from a different mitochondrial matrix protein, pyruvate dehydrogenase (Supplementary Fig. 3a–c and Supplementary Video 1), determining the rate of mtGFP accumulation in mitochondria by live time-lapse fluorescence imaging. The rate was significantly lower in ST-HdhQ111/111 than in ST-HdhQ7/Q7 cells (Supplementary Fig. 3c), supporting our findings obtained with the radiolabeled pOTC import assay using isolated mitochondria. Thus, two different assay systems using distinct mitochondrial presequences demonstrated that mutant Htt decreases mitochondrial protein import (Fig. 3a and Supplementary Fig. 3c).
Impaired protein import in HD synaptosomal mitochondria
Next we determined whether the mitochondrial protein import deficiency occurs in vivo in mutant Htt–expressing transgenic mouse brain. For these experiments, we used two lines of R6/2 mice, carrying 150 (±5) and 195 (±10) CAG repeats, respectively. The 150CAG R6/2 is the original R6/2 line18, whereas 195CAG R6/2 was spontaneously derived from the 150CAG R6/2 colony as a result of CAG repeat instability. Although the 195CAG R6/2 mice have a longer CAG repeat length, these mice die at 16–18 weeks of age, surviving approximately 3 weeks longer than 150CAG R6/2 mice. The investigation of two distinct R6/2 lines with differing CAG repeat lengths and disease severity may be informative, as it may reflect the heterogeneity of the human HD population.
To evaluate protein import activity in brain mitochondria, we used highly purified synaptosomal mitochondria from neuronal synapses and nonsynaptosomal mitochondria from all cell populations in the brain, including neuronal and non-neuronal cells (Fig. 3b). Because of the energetic demands of synaptic transmission, we hypothesized that synaptosomal mitochondria might be preferentially vulnerable to disturbances in mitochondrial function. We subjected nonsynaptosomal and synaptosomal mitochondria isolated simultaneously from forebrains of presymptomatic 150CAG R6/2 and control WT B6CBA mice at 22–24 d of age to the pOTC import assay (Fig. 3c). Nonsynaptosomal mitochondria from 150CAG R6/2 and WT mice did not exhibit a noticeable difference in protein import activity (Fig. 3c). In contrast, neuronal mitochondria purified from presymptomatic 150CAG R6/2 brain synaptosomes exhibited a 23 ± 6% (s.e.m.) reduction in protein import as compared to that of mitochondria from WT brains (Fig. 3c), suggesting that synaptic neuronal mitochondria may be more vulnerable to mutant Htt toxicity than nonsynaptosomal mitochondria. Similarly, synaptosomal mitochondria from 195CAG R6/2 mice in presymptomatic mice (5–6 weeks old) and those with mid-stage disease (10–11 weeks old) showed decreased protein import compared to that of mitochondria from WT brains (Fig. 3d). Nonsynaptosomal mitochondria isolated from 195CAG R6/2 mice 5–6 weeks old showed only a modest reduction in protein import compared to WT brains (Fig. 3d). Both 150CAG and 195CAG R6/2 synaptosomal mitochondria demonstrated a protein import defect early in disease pathogenesis, suggesting that this defect might act as an early trigger for synaptic mitochondrial dysfunction and subsequent neurodegeneration.
The clinical phenotype of HD is most selective to brain, and, in particular, to neurons. However, Htt is ubiquitously expressed in tissues outside of the CNS, including the liver. Expression of the N-terminal fragment of mutant human Httex1 in R6/2 mice is driven by the human HTT promoter, and thus R6/2 liver expresses mutant Htt. We therefore determined whether liver mitochondria isolated from presymptomatic 195CAG R6/2 mice exhibit abnormalities in protein import. Unlike brain synaptosomal mitochondria, liver mitochondria demonstrated little or no deficit in protein import in the presymptomatic (5–6 weeks) or middle stage of the disease (10–11 weeks), but did demonstrate a protein import deficit at end-stage disease (13–14 weeks) (Fig. 3e). This tissue difference in mitochondrial protein import activity between brain and liver suggests that the mitochondrial import defect is an early cellular event specific to brain mitochondria in HD and, in particular, to neuronal mitochondria.
HD import defect precedes mitochondrial respiratory dysfunction
The translocation of presequence proteins across the inner membrane to the matrix requires an intact mitochondrial membrane potential12. Therefore, it is possible that the import deficit in R6/2 mice results from an impairment of the mitochondrial respiratory chain, which generates the mitochondrial membrane potential.
To test this possibility, we assessed respiratory function in synaptosomal and nonsynaptosomal mitochondria isolated from forebrains of presymptomatic and mid-stage disease R6/2 mice at times when the import deficit in synaptosomal mitochondria is observed. We measured the resting (state 2) respiration rate with the addition of NAD-linked substrates, glutamate and malate, or FAD-linked substrate, succinate, using high-resolution respirometry and estimated the respiratory control ratio (Supplementary Fig. 4a,b). Overall, the respiratory function of synaptosomal and nonsynaptosomal mitochondria from presymptomatic 150CAG R6/2 mice and presymptomatic and mid-stage disease 195CAG R6/2 mice was not different from that of WT littermates (Supplementary Fig. 4a,b and data not shown). Thus, in two distinct R6/2 lines with two different CAG repeat lengths, brain mitochondria with a protein import deficit demonstrated preserved mitochondrial bioenergetic integrity. These results suggest that the impairment in protein import is not due to altered mitochondrial respiratory function or membrane potential, in accord with previous studies showing decreased respiratory chain function only in advanced disease in mice and in human HD19–21.
Impaired protein import in mutant Htt–expressing neurons
Because the protein import defect occurs in synaptosomal mitochondria from presymptomatic R6/2 mice before mitochondrial energetic failure, we hypothesized that import impairment may be an early, cell-intrinsic abnormality in neurons. To investigate mitochondrial protein import specifically in neurons before disease onset, we isolated mitochondria from primary cortical neurons prepared from embryonic day 15.5 (E15.5) 195CAG R6/2 and WT littermates and subjected them to the pOTC import assay (Supplementary Fig. 5a). Notably, we observed a modest but significant reduction in pOTC import in mitochondria isolated from R6/2 primary cortical neurons compared to those of WT littermates (Supplementary Fig. 5a), suggesting a mild neuron-specific import defect in HD mice.
HD is an age-dependent progressive neurodegenerative disease, and therefore age-related insults, including oxidative stress, may contribute to the progressive nature of this disease. We hypothesized that mitochondrial protein import may be specifically vulnerable to reactive oxygen species–induced stress in HD neurons as compared to normal neurons. To test this hypothesis, we assessed protein import activity in mitochondria prepared from WT and 195CAG R6/2 neurons exposed to a sublethal dose of hydrogen peroxide (10 µM), which does not affect mitochondrial protein import or survival in WT neurons (Fig. 4a and Supplementary Fig. 5b,c). Whereas sublethal hydrogen peroxide had little to no effect on import in WT neurons as compared to vehicle, hydrogen peroxide significantly decreased import in R6/2 neurons as compared to vehicle treatment (Fig. 4a), suggesting that mitochondrial protein import in R6/2 neurons is more vulnerable to sublethal oxidative stress, a phenomenon that may contribute to the age dependence observed in human HD.
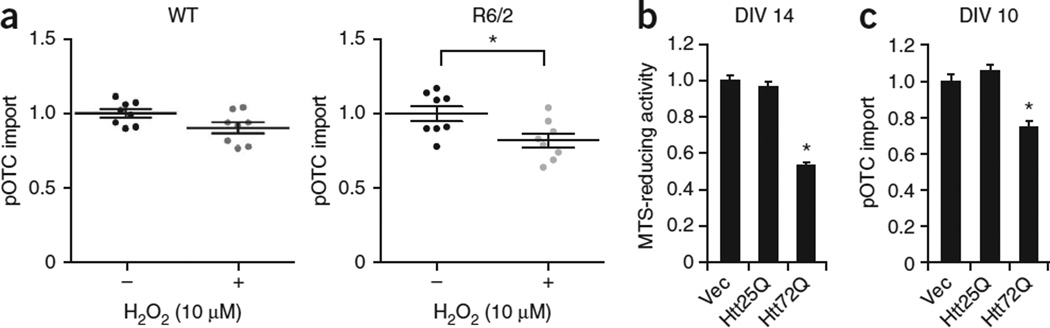
Figure 4.
Mutant Htt-expressing primary neurons show impaired mitochondrial protein import. (a) Primary cortical neurons from eight R6/2 and eight littermate WT embryos (E15.5) were individually plated into two dishes and treated at day in vitro (DIV) 7 with or without sublethal H2O2 (10 µM) for 2 h. Isolated mitochondria were then subjected to pOTC import assay (30 min). R6/2 neurons treated with sublethal H2O2 showed a significant decrease in pOTC import compared to vehicle-treated R6/2 neurons (unpaired t-test; *P = 0.022, t = 2.58, d.f. = 14, n = 8 cultures per condition prepared from 8 different embryos from 3 independent experiments). Data points are presented, with mean ± s.e.m. (b) Primary cortical neurons prepared from WT embryos were transduced with Httex1-25Q (Htt25Q), Htt72Q or control empty-vector lentivirus (Vec) at DIV 5. MTS assays were performed at DIV 14. Expression of Htt72Q, but not Htt25Q, decreased MTS-reducing activity, indicating that mutant Htt decreases mitochondrial metabolic activity (*P < 0.0001 compared to Htt25Q or vector control, n = 17 (vector), 12 (Htt25Q) or 12 (Htt72Q) cultures from 3 independent experiments). (c) Primary cortical neurons were transduced as in b. Mitochondria were isolated from neurons at DIV 10 before mutant Htt-induced neuronal death and subjected to pOTC import assays (30 min). Mitochondria isolated from neurons expressing Htt72Q exhibited decreased protein import compared to those expressing Htt25Q or vector control (*P < 0.0001, F2,18 = 24.42, n = 5 (vector), 10 (Htt25Q) or 6 (Htt72Q) mitochondria samples from 3 independent experiments). Htt25Q–expressing neurons showed no impairment of mitochondrial import compared to control neurons transduced with empty vector. Data (b,c) are presented as mean + s.e.m. One-way ANOVA, Bonferroni t-test.
We next took advantage of a primary cortical neuron model of HD to investigate the temporal relationships among mitochondrial protein import, mitochondrial metabolic activity and cell viability. In this system, lentivirally expressed mutant Httex1 (Httex1-72Q) but not WT Htt exon 1 (Httex1-25Q) accumulates as aggregates and decreases mitochondrial metabolic activity and cell viability (Fig. 4b and data not shown). We detected impaired protein import in mitochondria isolated from mutant Httex1-72Q–expressing neurons as compared to those expressing WT Httex1-25Q at a time before neurons begin to lose mitochondrial metabolic activity (Fig. 4c), suggesting that the mitochondrial protein import defect triggered by mutant Htt might cause cell death.
Because a pathological length of glutamine repeats is required for mutant Htt–induced impairment of protein import, we then asked whether mitochondrial import defect might be a common mechanism among various polyQ diseases. To address this question, we examined whether another polyQ disease protein, the mutant androgen receptor (AR), which is the causal protein in spinal and bulbar muscular atrophy (SBMA), would alter mitochondrial protein import in neurons by the pOTC import assay. Primary cortical neurons infected with mutant AR–expressing lentivirus (the N-terminal 127-amino-acid fragment of AR containing 65Q; AR-65Q) did not exhibit a decrease in mitochondrial protein import as compared to empty vector– or WT AR-22Q–infected neurons, whereas primary neurons expressing Httex1-72Q, but not Httex1-25Q, demonstrated diminished mitochondrial protein import (Supplementary Fig. 6a and data not shown), suggesting that inhibition of protein import is not a mechanism common to all polyQ diseases. In other experiments, expression of AR-65Q or Httex1-72Q, but not their WT counterparts, decreased mitochondrial metabolic activity in primary neurons (Supplementary Fig. 6b), suggesting that AR-65Q decreases neuronal viability by a mechanism independent of mitochondrial import.
Inhibition of protein import triggers neuronal death
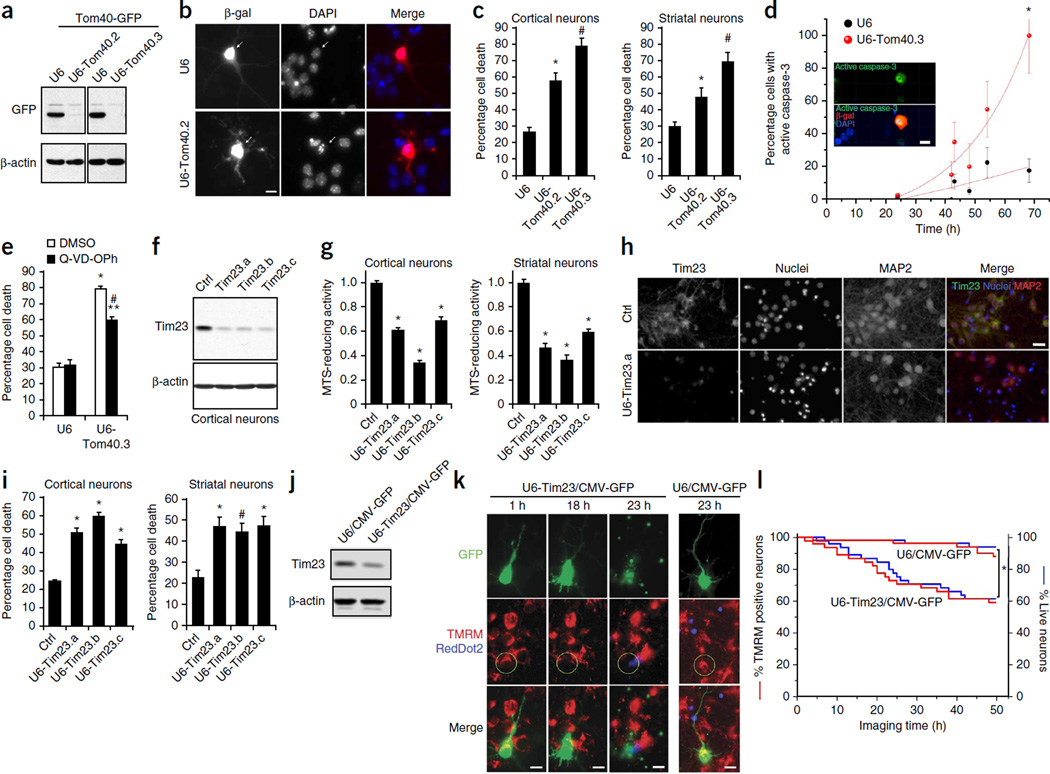
Given that protein import was perturbed in HD synaptosomal mitochondria early in the disease progression of R6/2 mice before mitochondrial bioenergetic failure (Fig. 3c,d and Supplementary Fig. 4a,b), in R6/2 primary cortical neurons (Fig. 4a and Supplementary Fig. 5a) and in mutant Htt lentivirus–infected primary cortical neurons before cell death (Fig. 4b,c), we hypothesized that impaired mitochondrial protein import might contribute to disease pathogenesis. To understand the biological consequences of deficient mitochondrial protein import in neurons, we first knocked down the Tom40 subunit, a core component of the TOM complex essential for import of almost all nuclearly encoded mitochondrial proteins, in primary neurons. Transfection of short hairpin RNA plasmids targeting two distinct regions of Tomm40, encoding Tom40, efficiently reduced the levels of exogenous and endogenous Tom40 in cell lines and primary neurons (Fig. 5a and Supplementary Fig. 7a,b). Mitochondria isolated from Tom40 knockdown cells demonstrated a decrease in protein import by 40% relative to that of control U6 plasmid–transfected cells (Supplementary Fig. 7c). Using these Tom40 shRNA plasmids, we then evaluated whether protein import was required for neuronal survival (Fig. 5b,c). Tom40 knockdown in primary cortical and striatal neurons triggered more cell death than vector transfection, suggesting mitochondrial protein import is essential for neuronal survival (Fig. 5b,c).
Figure 5.
Global and TIM23-driven mitochondrial protein import is necessary for survival of primary neurons. (a) Knockdown of Tom40 was confirmed using COS cells transfected with human Tom40-GFP expression plasmid and Tom40 RNAi (U6-Tom40.2, U6-Tom40.3) or control U6 plasmid. (b) DIV 5 cortical neurons were cotransfected with U6-Tom40.2 RNAi or U6 plasmid and with β-galactosidase (β-gal) expression plasmid and subjected to immunocytochemistry with anti–β-gal antibody at DIV 8. Arrows indicate representative β-gal+ control (top) and Tom40 RNAi (bottom) neurons, the latter of which show condensed or fragmented nuclei. (c) Neurons treated as in b were quantified for cell death by scoring nuclear morphology. Neuronal death was significantly increased in Tom40 knockdown cortical and striatal neurons compared to control neurons (one-way ANOVA, Fisher least significant difference; cortical neurons, *P = 0.0002, #P < 0.0001, F2,12 = 41.65, n = 5 coverslips from 4 independent experiments; striatal neurons, *P = 0.018, #P < 0.0001, F2,14= 19.46 ; n = 6 (U6 and U6-Tom40.3) or 5 (U6-Tom40.2) coverslips from 4 independent experiments). (d) Cortical neurons cotransfected with U6-Tom40.3 RNAi or U6 plasmid and with a β-gal plasmid were subjected to immunocytochemistry with anti–β-gal and anti-active caspase-3 antibodies and nuclear labeling with DAPI at the indicated time points after transfection. Active caspase-3+ neurons among β-gal+ transfected neurons were quantified. Inset: representative image of Tom40 knockdown neurons with active-caspase-3 immunoreactivity. Tom40 knockdown neurons showed increased active caspase-3 compared to control neurons (unpaired t-test; *P = 0.0005, t = 7.71, d.f. = 5, n = 6 slides per group from 3 independent experiments). (e) Transfected cortical neurons as in d were treated with Q-VD-OPh (20 µM) or vehicle (DMSO) and quantified for cell death as in c. qVD-OPh decreased Tom40 RNAi-induced cell death. *P < 0.0001 compared to U6 (DMSO), #P < 0.0001 compared to U6-Tom40.3 (DMSO), **P < 0.0001 compared to U6 (qVD-OPh), F3,20= 94.96, n = 6 coverslips per condition from 3 independent experiments. (f) Knockdown of Tim23 using shRNA lentiviruses (Tim23.a, Tim23.b and Tim23.c) was confirmed in cortical neurons by immunoblotting. Ctrl: luciferase shRNA lentivirus. (g) DIV 5 neurons infected with Tim23 shRNA or ctrl shRNA lentiviruses were subjected to MTS assays at DIV 12. Tim23 knockdown decreased mitochondrial metabolic activity in cortical and striatal neurons (cortical neurons: *P < 0.0001 compared to ctrl, F3,66 = 121.96, n = 13 (ctrl), 19 (Tim23.a, Tim23.b, Tim23.c) cultures per group from 6 independent experiments; striatal neurons: *P < 0.0001 compared to ctrl, F3,69 = 57.93, n = 16 (ctrl), 19 (Tim23.a, Tim23.b, Tim23.c) cultures per group from 6 independent experiments). (h,i) Neurons transduced as in g were fixed at DIV 12 and subjected to immunofluorescence with indicated antibodies and nuclear staining (Hoechst 33342). Cell death was assessed by nuclear morphology. Representative images of transduced cortical neurons (h). Tim23 knockdown significantly increased the percentage of dead cells in cortical (*P < 0.0001 compared to ctrl, F3,24= 60.98, n = 7 cultures per condition from 3 independent experiments) and striatal neurons (*P = 0.0004 and #P = 0.0013 compared to ctrl, F3,28= 7.623, n = 8 cultures per condition from 3 independent experiments) (i). (j) Knockdown of Tim23 was confirmed using N2a mouse neuroblastoma cells transfected with Tim23 RNAi plasmid (U6-Tim23/CMV-GFP) or control scrambled plasmid (U6/CMV-GFP) by immunoblotting. (k,l) DIV 5 cortical neurons were transfected with U6-Tim23/CMV-GFP or U6/CMV-GFP plasmid. At DIV 8, loss of mitochondrial membrane potential and cell death were assessed using TMRM and nuclear dye RedDot2 by live confocal imaging. Representative images (z projection) at indicated time points demonstrate Tim23 knockdown induces mitochondrial depolarization and subsequent cell death (k). The percentage of TMRM+ neurons and live neurons among GFP+ transfected neurons was quantified (l). Tim23 knockdown significantly decreased the number of TMRM+ neurons and live neurons compared to control transfection (log-rank test, *P = 0.002; n = 49 neurons from 3 wells per group). Experiments (a,f,j) were successfully repeated three times and full-length blots/gels are presented in Supplementary Figure 9. (c,e,i) 100–200 neurons per coverslip were counted. Scale bars: 10 µm (b,d,k), 20 µm (h). Data are presented as mean + s.e.m. (c,e,g,i) or mean ± s.d. (d). (e,g,i) One-way ANOVA, Bonferroni t-test.
Caspase activation has been demonstrated to function in cellular and in vivo models of HD22,23. We characterized Tom40 RNA interference– induced cell death in primary neurons by assessing the activation of caspase-3, an effector molecule of apoptotic signaling. Tom40 knockdown neurons demonstrated increased caspase-3 activation compared to that in vector-transfected neurons (Fig. 5d), and inhibition of caspase activation by a broad-spectrum caspase inhibitor, quinoline-Val-Asp-CH2-O-Ph (Q-VD-OPh), partially but significantly inhibited Tom40 RNAi-induced neuronal death (Fig. 5e). These results suggest that inhibition of mitochondrial protein import in neurons robustly activates caspase-dependent apoptotic signaling but that neuronal death occurs via caspase-dependent and caspase-independent pathways.
Given that the vast majority of nuclearly encoded mitochondrial proteins require the TOM import machinery for mitochondrial entry, our results indicate that global inhibition of mitochondrial protein import leads to neuronal death. Unlike the TOM complex, the TIM23 complex is responsible for the entry of a subset of, but not all, nuclearly encoded mitochondrial proteins. Our finding that mutant Htt binds to the TIM23 complex (Fig. 1c–g) raises the question of whether inhibition of TIM23-dependent import activity alone can also lead to mitochondrial dysfunction and neuronal death in neurons. Lentivirus-mediated delivery of shRNAs targeting three distinct regions of core TIM23 complex subunit mRNA Timm23, encoding Tim23, efficiently reduced the levels of endogenous Tim23 protein in primary neurons (Fig. 5f). Using these Tim23 shRNA lentiviruses, we evaluated whether Tim23 is required for mitochondrial function and cell survival in neurons. We subjected primary cortical and striatal neurons transduced with Tim23 RNAi lentiviruses to the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay to determine mitochondrial metabolic activity. Knockdown of Tim23 decreased MTS-reducing activity in neurons compared to that in control luciferase shRNA vector–infected neurons, suggesting that Tim23 is essential for mitochondrial metabolic activity (Fig. 5g). In addition, knockdown of Tim23 in primary cortical and striatal neurons triggered more cell death than control infection (Fig. 5h,i), suggesting that the TIM23 import complex is critical for neuronal survival.
To delineate the temporal relationship between mitochondrial dysfunction and death caused by Tim23 knockdown, we took advantage of live time-lapse imaging to examine the functional effects of Tim23 knockdown in primary neurons. Transfection of the Tim23 shRNA plasmid reduced the levels of endogenous Tim23 protein in cells (Fig. 5j). Following transfection, we assessed the loss of mitochondrial membrane potential and cell death by live confocal imaging in neurons using, respectively, TMRM, a fluorescent dye that accumulates only in polarized mitochondria, and RedDot2, a nuclear dye with high selectivity for membrane-compromised dead cells (Fig. 5k,l and Supplementary Video 2). Tim23 knockdown decreased the percentage of TMRM-positive neurons compared to that in control-transfected neurons, suggesting that defective TIM23-dependent protein import leads to loss of mitochondrial membrane potential (Fig. 5l). Notably, we found that Tim23 knockdown always induced loss of mitochondrial membrane potential before cell death (by 3.4 ± 0.13 h; n = 25 U6-Tim23/CMV-GFP neurons) (Fig. 5k,1 and Supplementary Video 2). Together, our results indicate that impairment of both global and TIM23-driven mitochondrial protein import leads to neuronal death.
Deficient protein import contributes to neuronal death by mutant Htt
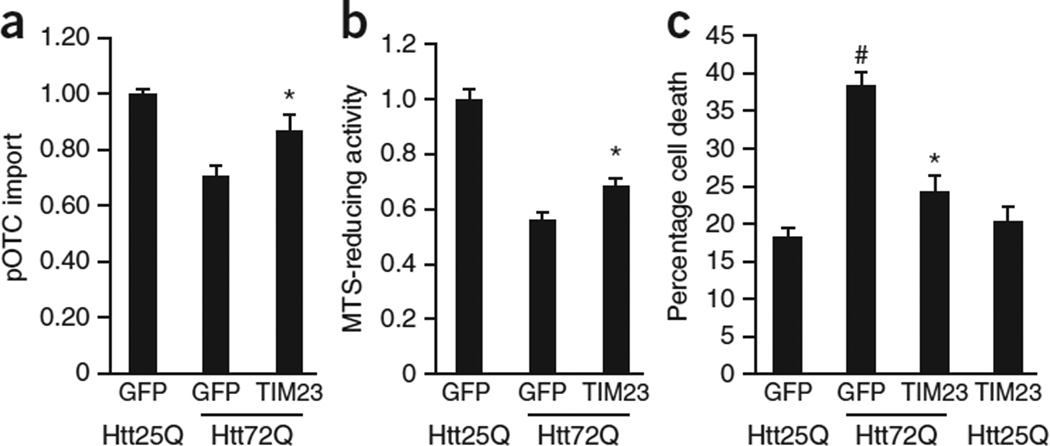
The finding that protein import was impaired in mutant Htt–expressing primary neurons before decreased mitochondrial metabolic activity (Fig. 4b,c) raises the hypothesis that impaired import may drive cell death. We therefore increased protein import in mutant Httex1-72Q–expressing neurons through enforced expression of the TIM23 complex by lentivirus-mediated expression of the three major subunits, Tim23, Tim50 and Tim17a (Fig. 6a). We confirmed the expression of these subunits by quantitative reverse transcription (qRT)-PCR and by immunoblotting (data not shown).
Figure 6.
Augmentation of mitochondrial protein import rescues neurons from mutant Htt-induced death. (a) DIV 5 cortical neurons were transduced with WT Httex1-25Q (Htt25Q) or mutant Httex1-72Q (Htt72Q) lentivirus and at DIV 6 were cotransduced with lentivirus expressing three subunits of the TIM23 complex, Tim23, Tim50 and Tim17a. Mitochondria isolated from transduced neurons at DIV 10 were subjected to pOTC import assay (30 min). Overexpression of the TIM23 complex subunits increased pOTC import in Htt72Q neurons (*P = 0.007 compared to GFP-expressing Htt72Q neurons, F2,27 = 14.05, n = 10 samples per condition from 5 independent experiments). (b) Primary cortical neurons transduced as in a were subjected to MTS assays at DIV 14. Overexpression of the TIM23 complex subunits partially but significantly metabolic activity (*P = 0.006 compared to GFP-expressing Htt72Q neurons, F2,45 = 59.39, n = 17 wells per group for GFP-expressing Htt25Q and GFP-expressing Htt72Q neurons, n = 14 wells for TIM23-expressing Htt72Q neurons from 4 independent experiments). (c) Primary cortical neurons transduced as in a were assessed for cell death by scoring nuclear morphology at DIV 14. Htt72Q–expressing neurons showed increased cell death compared to Htt25Q–expressing neurons (#P < 0.0001, GFP-expressing Htt72Q compared to GFP-expressing Htt25Q neurons). Overexpression of the TIM23 complex subunits in Htt72Q neurons inhibited neuronal death (*P < 0.0001 compared to GFP-expressing Htt72Q neurons). F3,51 = 29.47, n = 16 (GFP-expressing Htt25Q neurons), 15 (GFP-expressing Htt72Q neurons) and 12 (TIM23-expressing Htt72Q and TIM23-expressing Htt25Q neurons) wells per group from 3 independent experiments; 200 neurons were counted per well. (a–c) Data are presented as mean + s.e.m. One-way ANOVA, Bonferroni t-test.
Overexpression of the major subunits of the TIM23 complex in mutant Httex1–expressing primary neurons rescued the mitochondrial protein import defect and partially but significantly increased mitochondrial metabolic activity (Fig. 6b). Notably, overexpression of the TIM23 complex subunits inhibited mutant Htt–induced cell death (Fig. 6c). Taken together, these findings indicate that mutant Htt–induced inhibition of mitochondrial protein import can cause mitochondrial dysfunction and neuronal death. Collectively, we have provided a mechanism by which mutant Htt directly impairs mitochondrial protein import through an interaction with the import machinery and have demonstrated that this mitochondrial import defect triggers mutant Htt–induced cell death (Supplementary Fig. 8).
DISCUSSION
Direct association of N-terminal mutant Htt fragments with mitochondria has been found in transfected primary neurons and brain neurons from several HD transgenic mice9–11. In this study, we have demonstrated that mutant Htt localizes to mitochondria in the caudate nucleus of patients with HD, suggesting a direct toxic effect of mutant Htt on mitochondria. Although a large body of evidence has implicated mitochondrial dysfunction as central to HD, the underlying molecular basis for mutant Htt–induced mitochondrial abnormalities had remained largely undefined. We have found that mutant Htt interacts with the mitochondrial TIM23 translocase complex and directly causes protein import deficiency. We also provide evidence that mutant Htt–induced protein import defect causes mitochondrial dysfunction and neuronal death in primary neurons. The machinery provides not only a specific molecular basis for the association of mutant Htt with mitochondria but also reveals an important biological process disrupted by mutant Htt, leading to mitochondrial dysfunction. Thus, our study highlights an important and early contribution of disruption of the mitochondrial protein import system to neuronal death in HD.
Several mitochondrial deficits, including decreased membrane potential and respiratory function, decreased calcium buffering capacity, and altered mitochondrial number and morphology, have been associated with HD3,11,16,24–27. The defect in protein import is an early, specific event observed in R6/2 forebrain neuronal mitochondria, as we found no alterations in respiratory function using the respiratory complex I and II substrates at a time when import deficits are observed. These results suggest that protein import deficiency is not a consequence of a reduction in mitochondrial membrane potential and likely drives further mitochondrial insults by decreasing the levels of key imported proteins involved in essential mitochondrial functions. Reduced import of proteins involved in the tricarboxylic acid cycle, oxidative phosphorylation, defense mechanisms against reactive oxygen species, and mitochondrial DNA repair may result in lower mitochondrial ATP production and increased oxidative damage of mitochondrial proteins and DNA, which are observed in HD9,28–31. Previous studies using isolated mitochondria showed that the N-terminal fragment of mutant Htt or expanded polyQ domain alone directly decreases the calcium threshold for the mitochondrial permeability transition pore, calcium retention capacity and mitochondrial membrane potential16,17,24. The relationship between these mitochondrial deficits and protein import will be a worthwhile focus for future studies.
Mitochondrial morphology is regulated by a dynamic balance between fusion and fission. Recent studies have suggested that mutant Htt affects mitochondrial dynamics via its abnormal interaction with and activation of the mitochondrial fission GTPase DRP1 to increase mitochondrial fragmentation11,27,32. Notably, a previous yeast mutant screen demonstrated that yeast lacking subunits of the TOM, the sorting and assembly machinery (SAM) or the TIM23 complexes show severe defects in mitochondrial morphology, suggesting that mitochondrial protein import, assembly and sorting are required for mitochondrial morphogenesis33,34. Thus, it is conceivable that the early defect in mitochondrial protein import in neurons of presymptomatic HD-affected brain might lead to aberrant mitochondrial morphology by affecting the levels of proteins related to mitochondrial morphogenesis.
A fundamental question in HD is why the brain and, in particular, neurons are selectively affected, even though mutant Htt is expressed throughout the body. The early impairment in mitochondrial protein import observed in forebrain synaptosomal mitochondria of R6/2 mice but not liver mitochondria suggests that the mitochondrial protein import defect occurs in a tissue-, cell type– and time-dependent manner. To our knowledge, this is the first time a mitochondrial protein import defect has been observed in synaptosomal mitochondria in any neurological disease. It is noteworthy that, in HD knock-in mouse brain, ATP levels are reduced in the synaptosomal fraction, but not in the cytoplasmic fraction, late in disease9. Together, these findings suggest the decreased levels of ATP and degenerated mitochondria in synaptic sites in HD may result from an early impairment in mitochondrial protein import. Given that presynaptic mitochondria are important for synaptic vesicle release and recycling, it is possible that the early protein import deficit in synaptic mitochondria in HD model mice may result in altered synaptic transmission, leading potentially to the neuronal dysfunction observed in patients with HD35.
Mitochondrial protein import in primary R6/2 cortical neurons was particularly sensitive to sublethal hydrogen peroxide compared to that in WT neurons. Oxidative stress may thus amplify a neuronal vulnerability in mitochondrial protein import over time and contribute to the progressive deficits seen in patients with HD. Because antioxidants have been shown to be effective in slowing disease progression in HD transgenic mice36, our results raise the possibility that the therapeutic effect of antioxidants may be mediated, at least in part, by modifying mitochondrial protein import.
The mechanism of mutant Htt action on mitochondrial protein import and the downstream mitochondrial pathophysiology has implications for diverse neurological disorders. Mitochondrial protein import has been implicated in other neurodegenerative diseases, including Alzheimer’s disease and amyotrophic lateral sclerosis37–39. Interestingly, genetic mutation of an import-associated gene has been shown to lead to the X-linked recessive disorder Mohr-Tranebjaerg syndrome (also known as deafness dystonia syndrome), a progressive neurodegenerative disease caused by mutations in the deafness-dystonia protein 1 (DDP1) gene, TIMM8A (ref. 40). A link between an import defect and neurological disease has also been demonstrated in heterozygous Timm23 knockout mice, which exhibit a neurological phenotype, although the exact molecular and cellular nature of the abnormality remains to be elucidated41. Our finding that a mitochondrial import deficit directly causes neurodegeneration raises the possibility that other neurodegenerative diseases exhibiting import defects might use common cell death mechanisms. Impairment of mitochondrial protein import in neurons triggered cell death by caspase-dependent and caspase-independent mechanisms, consistent with reported mechanisms of mutant Htt–induced cell death observed in striatal cell lines and in mice in vivo22,23. Recent studies have identified proteins that monitor mitochondrial homeostasis through the protein import machinery42, raising the possibility that such molecular sensors may activate specific death pathways in response to a global decrease in protein import. The identification of specific matrix proteins whose levels are decreased in mitochondria in HD neurons may also provide insights into the detailed mechanism of how protein import inhibition might lead to neuronal death.
In addition to HD, there are eight polyQ diseases, including SBMA, dentatorubral-pallidoluysian atrophy and spinocerebellar ataxia type 1, that are caused by the expansion of a polyQ tract in specific proteins and are also progressive neurodegenerative disorders with the loss of selective neurons43,44, raising the possibility that other polyQ diseases may trigger protein import defects. Our experiments, however, indicate that the causal protein in SBMA, mutant AR, which harbors an expanded glutamine repeat, has little to no effect on mitochondrial protein import, suggesting that import deficits are not common to all polyQ disorders, consistent with the requirement for both the N17 N-terminal sequence and the polyQ of mutant Httex1 for interaction with the TIM23 complex. It will be of great future interest to examine whether other polyQ disease proteins might carry an N17-like sequence and therefore potentially affect mitochondrial protein import.
In addition to a direct effect of mutant Htt on mitochondrial function, many lines of evidence have suggested that transcriptional dysregulation is important to the mechanism of mitochondrial dysfunction. PGC-1α, a key transcriptional coactivator that regulates the expression of genes involved in energy metabolism and mitochondrial biogenesis, has been shown to be downregulated in HD25,45,46. As such, therapeutic strategies targeting both the direct action of mutant Htt on mitochondria and the transcriptional deregulation of mitochondrial proteins may thus be necessary to effectively rescue mitochondrial dysfunction in HD.
ONLINE METHODS
HD transgenic R6/2 mice
R6/2 mice, which carry the promoter sequence and exon 1 of a mutant human HTT gene with approximately 150 CAG repeats, were obtained from JAX (Bar Harbor, ME). A colony was maintained by breeding R6/2 males with B6CBAF1 females (JAX). R6/2 mice with a spontaneous expansion of CAG repeats, 195 ± 10 CAG repeats, were found in the colony and further maintained. PCR genotyping was performed using a primer set (CGGCTGAGGCAGCAGCGGCTGT and GCAGCAGCAGCAGCAACAGCC GCCACCGCC) as described18. To maintain mice carrying the same number of CAG repeats, a second PCR analysis was also conducted using a primer set amplifying across the CAG repeats (ATGAAGGCCTTCGAGTCCCTCAAGTCCTTC and GGCGGCTGAGGAAGCTGAGGA). The precise CAG repeat length in the R6/2 mice was determined by ABI377 sequencer using tail DNA (Laragen Inc., Culver City, CA). All live vertebrate experiments were performed in compliance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals. Animal protocols were approved by the Institutional Animal Care and Use Committees of the University of Pittsburgh and Washington University.
Antibodies
Rabbit polyclonal anti-Tom40 (H-300; sc-11414, Santa Cruz Biotechnology; 1:300 dilution for immunoblotting), goat polyclonal anti-Tim17a (sc-13293, Santa Cruz Biotechnology; 1:200 for immunoblotting), rabbit polyclonal anti-Tim23 (ab116329, Abcam; 1:200 for immunofluorescence), rabbit polyclonal anti-DRP1 (ab54038, Abcam; 1:200 for immunofluorescence), rabbit monoclonal anti-Tim50 (ab109436, Abcam; 1:2,000 for immunoblotting), mouse monoclonal anti-Tim23 (611222, BD Transduction Laboratories; 1:2,000 for immunoblotting), mouse monoclonal anti–β-galactosidase (β-gal) (Z378B, Promega; 1:600 for immunofluorescence) and mouse monoclonal anti–β-actin (A5411, Sigma; 1:5,000 for immunoblotting) antibodies were purchased. Anti–cleaved caspase-3 (Asp175) antibody (9661, Cell Signaling Technology; 1:1,000 dilution for immunofluorescence), specifically recognizing active caspase-3, and mouse monoclonal anti-Htt antibody EM48 (MAB5374, Millipore; 1:200 dilution for immunofluorescence), generated against the first 256 amino acids of human Htt with a deletion of the polyQ tract, were purchased. Mouse monoclonal anti-Htt (MAB5492, Millipore; 1:1,000 dilution for immunoblotting), mouse monoclonal anti–expanded polyglutamine (clone 1C2; MAB1574, Millipore; 1:1,000 dilution for immunoblotting and immunofluorescence) and rabbit polyclonal anti-GFP (A6455, Life Technologies (Molecular Probes); 1: 2,000 dilution for immunoblotting) antibodies were purchased.
DNA constructs
A bacterial expression mutant Htt construct encoding human mutant HTT exon 1 with 97 polyglutamine repeats, pGEX4T3-Httex1-97Q, was generated by standard subcloning. The bacterial expression construct for GST-Httex1–23Q (ref. 24) was kindly provided by M. Lesort (University of Alabama, Birmingham, AL). GFP-tagged human N170 WT and mutant Htt expression plasmids were provided by M.E. MacDonald (Harvard Medical School, Boston, MA). Tim23 and Tom40 shRNA plasmids were produced by insertion of annealed oligonucleotides containing the following targeted sequences into pBSU6/CMV-GFP and pBSU6, respectively: pBSU6-Tim23/CMV-GFP: CGGAGGAAGTAGCAACAAA; pBSU6-Tom40.1: GGAGTGCCACCGGAAGTGCAA; pBSU6-Tom40.2: GCTGAGTCCCAC AGAGGCGTT; pBSU6-Tom40.3: GGCACTGTCATGTCTCTAGCT. All constructs were confirmed by sequencing. Lentivirus-based Tim23 RNAi constructs (pLKO.1-puro), developed at the Broad Institute of MIT and Harvard, were obtained (RNAi Core, Washington University). The Tim23 targeted sequences are as follows: pLKO.1-Tim23.a: GCTGTGACAAAGATCATGGAT; pLKO.1-Tim23.b: GCCTGGTCCAAACCAAGAAAT; pLKO.1-Tim23.c: CGGTCTTCGTTT AGGATTGAA. Tom40-GFP expression plasmid47 was kindly provided by M. Ryan (La Trobe University, Melbourne, Australia). Mouse Timm23, Timm50 and Timm17a cDNAs were cloned by reverse transcription–PCR from mouse brain RNA and subcloned into lentiviral expression plasmid pRRLsinPGK (Hope Center Viral Vectors Core, Washington University). Lentiviral expression plasmids containing Httex1–25Q and Httex1–72Q constructs under the control of the mouse PGK (Pgk1) promoter (mPGK-Httex1-25Q and mPGK-Httex1-72Q) were kindly provided by D. Krainc (Harvard Medical School, Boston, MA). cDNA clones encoding the N-terminal 127-amino acid fragment of androgen receptor (AR) containing 22Q and 65Q were kindly provided by M. Diamond (Washington University, St. Louis, MO) and were subcloned into the mPGK lentiviral expression vector.
Cell culture, DNA transfection and lentiviral transduction
Mouse primary cortical and striatal neurons from embryonic day (E) 15.5 Swiss Webster mouse fetuses were cultured in plating medium (minimal essential medium (MEM), 10% FBS, 0.45% glucose, 1 mM sodium pyruvate, 2 mM glutamine, 20 U/ml penicillin and 20 µg/ml streptomycin for 3 h and then maintained in serum-free Neurobasal medium containing B27 supplement (Life Technologies), 0.5 mM glutamine and 25 µM glutamate for the first 3 d in a humidified incubator (37 °C in 5% CO2). Half of the medium was replaced with Neurobasal medium with B27 and 0.5 mM glutamine every 3 d. At 5 d in vitro (DIV 5), neurons were transfected with pBSU6-GFP plasmids or cotransfected with pBSU6 RNAi and β-gal plasmids using Lipofectamine 2000 (Life Technologies). Primary cortical and striatal neurons were infected with Tim23 RNAi lentiviruses at DIV 5 and subjected to mitochondrial metabolic activity assays using MTS (Promega) or cell death assays at DIV 12. For the immunoblotting experiments evaluating Tim23 knockdown efficiency in transduced neurons, neurons were harvested at DIV 10. For the rescue experiments, mouse primary cortical neurons plated on 6-cm or 96-well plates were infected with lentiviruses expressing Httex1-25Q, Httex1-72Q or control empty vector at DIV 5. On the next day (DIV 6), the same neurons were infected with Tim23, Tim17a and Tim50 lentiviruses or GFP lentivirus as control. Viral copy number was adjusted for transduction of neurons on the basis of titer measured using the Lenti-X qRT-PCR titration kit (Clontech). Neurons on 6-cm plates were harvested for mitochondrial protein import assay at DIV 10; neurons on 96-well plates were subjected to MTS assay or cell death assay at DIV 14.
R6/2 and littermate WT primary cortical neurons were prepared from E15.5 embryos, which were obtained from pregnant WT B6CBA females mated with a 195CAG R6/2 male. Neurons obtained from each embryo were cultured separately on poly-l-lysine–coated plates and subjected to in vitro pOTC import assay on DIV 7–8. The genotype of cultured neurons was determined by PCR using embryonic tail DNA.
Mouse knock-in striatal cell lines, ST-HdhQ111/Q111 and ST-HdhQ7/Q7 (ref. 48), were obtained from M.E. MacDonald (Harvard Medical School, Boston, MA) and were cultured at 33 °C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS and 1 mM sodium pyruvate. The immortalized rat striatal cell line ST14A (ref. 49), as well as ST14A stably expressing the N-terminal 548 amino acid fragments of WT or mutant Htt, which were obtained from E. Cattaneo (University of Milan, Milan, Italy), were cultured at 33 °C in 5% CO2 in DMEM containing 10% FBS and 0.1 mM nonessential amino acids (Life Technologies). These striatal cell lines stably express the temperature-sensitive SV40 large T antigen and stop dividing when cultured at the nonpermissive temperature of 37 °C. Cells were cultured at 37 °C for 24–36 h before mitochondrial isolation for in vitro pOTC import assays. Human embryonic kidney (HEK) 293 cells were cultured at 37 °C in 5% CO2 in DMEM containing 10% FBS. For transient transfection experiments with cell lines, cells were grown on poly-l-lysine coated plates and transfected with indicated plasmids using Lipofectamine 2000 (Life Technologies).
Mitochondria-targeted GFP (mtGFP), which contains a presequence from E1-a pyruvate dehydrogenase, was expressed in ST-Hdh cells using the BacMam system (Life Technologies).
Cell death assays
Primary neurons infected with Tim23 shRNA or control luciferase shRNA lentivirus at DIV 5 were fixed 7 d after infection. Nuclei were stained with Hoechst 33342, and neurons were assessed in a blinded fashion for cell death by scoring condensed or fragmented nuclei. Primary neurons plated in a 24-well plate were transfected with Tom40 shRNA or control pBSU6 plasmid along with β-gal expression plasmid at DIV 5, fixed 3 d after transfection and subjected to indirect immunofluorescence with anti–β-gal antibody and nuclear DAPI staining. β-gal–positive neurons were assessed for cell death as described above. Experiments were performed in duplicate or triplicate in three or more independent experiments.
Isolation of mitochondria
Brain synaptosomal and nonsynaptosomal mitochondria were isolated from R6/2 mice and sex-matched littermate or sex- and age-matched WT mice using differential centrifugation followed by discontinuous Percoll gradient centrifugations as described50. Briefly, 2–3 mouse forebrains were homogenized in IM buffer (5 mM HEPES-Tris (pH 7.4), 225 mM sucrose, 75 mM mannitol and 1 mM EGTA) and then centrifuged at 1,300g for 3 min. The supernatant was spun at 12,700g for 10 min. The resulting pellet was resuspended in 15% Percoll, laid on top of 24% and 40% Percoll, and subjected to centrifugation at 30,700g for 8 min. The band at the boundary between 24% and 40% Percoll contains nonsynaptosomal mitochondria and the band in the 24% Percoll contains the fraction enriched in synaptosomes. The nonsynaptosomal mitochondrial fraction was collected, washed with IM buffer to remove Percoll, spun down and resuspended in the IM buffer without EGTA. The synaptosomal fraction was diluted two times with IM buffer and placed in the nitrogen disruption vessel (45 ml; Parr Instrument, cat. no. 4639) and incubated on ice for 15 min at 1,500 p.s.i. The disrupted synaptosomal fraction was layered on top of 24% Percoll and centrifuged at 30,700g for 8 min. The bottom fraction enriched with synaptosomal mitochondria was collected, washed with IM buffer, spun down and resuspended in IM buffer without EGTA. Isolated mitochondria were kept on ice and used for mitochondrial protein import assay and/or respiratory function assays within 3 h after preparation. Liver mitochondria were isolated as described previously51. Mitochondria from primary cultured neurons or cell lines were isolated as described with a modification to the homogenization step52. Cells were homogenized in mitochondrial isolation buffer by passing through a 23-G needle 15–20 times, and the homogenates were subjected to differential centrifugation (600g and 8,000g) to obtain the mitochondrial fraction.
Preparation of GST-Htt exon 1 fusion proteins from bacteria
GST and GST-Htt exon 1 (GST-Httex1) fusion proteins were purified from transformed BL21star (DE3) cells (Life Technologies) as previously described24. GST fusion proteins bound to glutathione 4B Sepharose beads (GE Healthcare) were eluted with 50 mM Tris buffer (pH 8) containing 10 mM reduced glutathione, and the purified proteins in the glutathione elution buffer were concentrated in PBS using AmiconUltra-10K (Millipore).
GST pull-down assays and protein sequence analysis by LC-MS/MS
Mitochondria isolated from adult mouse forebrains were incubated with GST, GST-Httex1-23Q or GST-Httex1-97Q in the mitochondrial isolation buffer (3 mM HEPES-KOH, pH 7.6, 210 mM mannitol, 70 mM sucrose and 0.2 mM EGTA) for 1 h on ice and then lysed in TNE buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% NP-40) containing protease inhibitors on ice for 30 min. Extracts were spun at 21,130g at 4 °C, and clarified supernatants were incubated with glutathione 4B Sepharose beads (GE Healthcare) at 4 °C overnight. The glutathione beads were then washed extensively with ice-cold TNE buffer, and bound proteins were subjected to SDS-PAGE followed by immunoblotting analysis. For the samples subjected to mass spectrometry, glutathione bead–bound proteins from equal amounts (600 µg) of mitochondria were further washed with high stringency RIPA buffer following TNE buffer wash to decrease nonspecific interaction. Proteins were then eluted from the glutathione beads in SDS sample buffer, separated by SDS-PAGE and stained with Coomassie. Proteins in the gel were digested with trypsin and analyzed by mass spectrometry (LC-MS/MS) at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School, Boston, MA).
Immunoprecipitation
Cell or brain lysates were prepared in TNE buffer containing protease inhibitors and phosphatase inhibitors and immunoprecipitated with anti-Tim50 rabbit monoclonal antibody followed by protein A magnetic beads (Life Technologies). The immunoprecipitates were subjected to SDS-PAGE and immunoblotting with indicated antibodies, and the proteins were visualized using enhanced chemiluminescence (Pierce).
Mitochondrial poTc import assay
The assay was performed as previously described52,53. Human ornithine transcarbamylase (OTC) precursor cDNA in pGEM-3Zf(+)-pOTC plasmid, which was kindly provided by M. Yano (Kumamoto University, Kumamoto, Japan), was transcribed and translated in vitro using the TNT-coupled reticulocyte lysate system (Promega) in the presence of l-[35S]methionine (PerkinElmer). Following translation, [35S]methionine-labeled pOTC was incubated with isolated mitochondria at 25 °C for the indicated times, and mitochondria containing imported OTC were collected by centrifugation (9,000g, 10 min) and subjected to SDS-PAGE. The radioactive polypeptides on the gel were visualized by fluorography with Amplify (GE Healthcare) followed by exposure to X-ray film. Cleaved mature OTC (mOTC), which represents the completion of import into the mitochondrial matrix and migrates faster than pOTC on SDS-PAGE, was quantified by ImageJ (NIH). The data are presented as the percentage of mOTC compared to input (total [35S]pOTC amount added to each reaction) and scaled to mOTC in control mitochondria after the maximum reaction time (set equal to 1) unless otherwise specified. In the import assay with forebrain mitochondria, data are scaled to mOTC in control WT mitochondria at 60 min reaction time (set equal to 1).
Mitochondrial respiration measurements
Respiration in isolated mitochondria was measured in buffer containing 125 mM KCl, 5 mM HEPES-KOH (pH 7.4), 2 mM KH2PO4, and 5 mM glutamate and malate or 10 mM succinate as respiratory substrates using the Oroboros 5 mM Oxygraph-2k high-resolution respirometry system (Oroboros, Austria) equipped with two Clark-type electrodes. Respiration in the presence of substrates-only corresponds to resting state (state 2) respiration. The subsequent addition of ADP (100 µM) initiates ATP synthesis coupled to proton reentry across the membrane, which corresponds to ADP-stimulated (state 3) respiration. ADP exhaustion then leads to a reduction of the respiratory rate and corresponds to state 4 respiration. The respiratory control ratios (RCR), an index of the efficiency of coupled respiration to phosphorylate ADP, were calculated as the ratios of state 3 to state 2 respiration rates (RCR3/2) or the ratios of state 3 to state 4 respiration rates (RCR3/4) for both respiratory complex I– and complex II–dependent respiration using glutamate-malate and succinate, respectively.
Immunofluorescence microscopy
Cells were fixed in 4% paraformaldehyde in PBS for 20 min, permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature and subjected to immunofluorescence with the indicated primary antibodies and secondary antibodies, Cy3-conjugated goat anti-mouse IgG (115-165-146, Jackson ImmunoResearch, 1:300 dilution) and/or Alexa Fluor 488–conjugated goat anti–rabbit IgG (A11034, Life Technologies (Molecular Probes); 1:300) antibodies, according to standard protocols. Cell nuclei were labeled with DAPI or Hoechst 33342. Images were captured using SensiCam CCD camera with imaging software IPLab 4.0 (BD Biosciences) through a fluorescence microscope (Nikon) or using a confocal laser scanning microscope system (FluoView FV1000; Olympus, Inc.).
For immunostaining of human grade-2 HD and control age-matched caudate nucleus samples, sections (50 µm) were incubated with indicated antibodies in Tris buffer (pH 7) containing 0.3% Triton X-100 for 24–72 h at 4 °C, followed by incubation with FITC-conjugated horse anti–mouse IgG (FI-2001, Vector, 1:200) and Cy3-congugated donkey anti–rabbit IgG (711-165-152, Jackson ImmunoResearch; 1:200) antibodies. Digital imaging was performed using an IX81 microscope (Olympus, Inc.) equipped with an IX2-DSUA-SP confocal spinning disk and a 100× UPLSAPO objective (NA 1.40; Olympus, Inc.) and UIS dichromatic mirror and emission filter sets (Olympus, Inc.). Images were captured with a cooled charge-coupled device camera (Orca R2; Hamamatsu). Optical sections and three-dimensional image reconstructions were performed using Metamorph software (Molecular Devices, LLC). Images were captured at 0.2-µm intervals for 50 focal planes, and stacks were deconvolved with a constrained iterative algorithm.
Time-lapse live cell imaging
Mouse primary cortical neurons were plated at 0.3 × 106 cells per well of 24-well #1.5-coverglass-bottom plates (In Vitro Scientific) and transfected with a Tim23 RNAi or control pBSU6-GFP plasmid at DIV 5. Three days after transfection, neurons were loaded with the red fluorescent TMRM dye (200 nM) and the cell membrane–impermeable, far-red nuclear dye RedDot2 and placed into an on-stage incubation chamber (WSKM-FI; Prior Scientific, Rockland, MA) at 37 °C and 5% CO2. Sets of images were acquired every 1.5 h using an Olympus IX81-DSU inverted confocal microscope with UPLSAPO 40× air 0.95 NA lens, Lumen 200 (Prior Scientific) light source, H117 motorized linear encoded x-y stage (Prior Scientific) and Hamamatsu Orca R2 CCD camera, and analyzed using Metamorph image acquisition software (Molecular Devices). For quantitative analyses, GFP-positive transfected neurons were assessed over time for the loss of TMRM signal from mitochondria and the appearance of RedDot2 signal in the nucleus, which represent mitochondrial depolarization and cell death, respectively. Data collection and analysis were performed in a manner blinded to the conditions of the experiments.
Human brain tissues
Postmortem striatal tissue specimens from three patients with neuropathological grade 2 HD (sex, age (years): male, 54; male, 46; female, 68) and three controls (female, 67; female, 57; male, 44) were subjected to immunohistochemical analysis. The brain specimens were received from the Bedford Veterans Administration Medical Center Brain Tissue Archive. The postmortem intervals did not exceed 15 h and were similar between controls and HD patients. Work involving human brain tissue samples was approved by the IRB and the Committee for Oversight of Research Involving the Dead at the University of Pittsburgh.
Statistics
Statistical analyses were performed with Prism 6 and XLSTAT2012 software. Data are obtained from at least three independent experiments and expressed as mean ± s.e.m. unless otherwise specified. The Student’s t-test (unpaired, two-tailed) for parametric data and the Mann-Whitney U test for nonparametric data were used for analysis of two groups. Equal variance for parametric data was formally tested using an F-test. In experiments with more than two groups, analysis of variance (ANOVA) was performed followed by Fisher’s least significant difference (three groups) or Bonferroni test (three groups or greater) for pairwise comparisons. For time-lapse imaging analysis, Kaplan-Meier curves were used to estimate survival function, and statistical comparisons between Tim23 knockdown and control neurons were made using the log-rank test. No randomization was used, but treatments and assays for different conditions were performed in a blinded fashion. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications54.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to J.T. Greenamyre, B. Kristal, I.G. Stavrovskaya and members of the Friedlander laboratory for discussion and technical support. We thank M. Lesort for assistance generating recombinant GST-Httex1. This work was supported by US National Institutes of Health grants R01 NS039324 and NS077748 (to R.M.F.) and K01 AG033724 (to H.Y.), Huntington’s Disease Society of America (Coalition for the Cure) (R.M.F.), the Brain & Behavior Research Foundation (NARSAD Young Investigator Award) (H.Y.) and the DSF Charitable Foundation (R.M.F.).
Footnotes
Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
H.Y. designed and performed experiments, analyzed data and wrote the manuscript. S.V.B. contributed to brain mitochondrial preparation, functional and imaging analysis of mitochondria, and manuscript writing. O.V.B. performed preparation of GST fusion proteins and primary neuron cultures and assisted in experiments designed by S.V.B. and H.Y. J.K. and R.J.F. performed immunofluorescence experiments with human HD samples and cell lines. Y.P. performed viral transduction of primary neurons and assisted with neuron viability assays, preparation of GST fusion proteins and coimmunoprecipitation experiments. S.Y. performed Tim23 knockdown experiments for live imaging. D.L.C. contributed to the design of experiments. A.H.K. contributed to neuronal death experiments, experimental design and manuscript writing. R.M.F. contributed to the development of the project, supervised the analysis of all experiments and contributed to manuscript writing.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hersch SM, Rosas HR, Ferrante RJ. Neuropathology and pathophysiology of Huntington’s disease. In: Watts RL, Standaert DG, Obeso JA, editors. Movement Disorders. 3rd edn. New York: McGraw-Hill; 2012. p. 683. [Google Scholar]
- 2.The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Damiano M, Galvan L, Deglon N, Brouillet E. Mitochondria in Huntington’s disease. Biochim. Biophys. Acta. 2010;1802:52–61. doi: 10.1016/j.bbadis.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Costa V, Scorrano L. Shaping the role of mitochondria in the pathogenesis of Huntington’s disease. EMBO J. 2012;31:1853–1864. doi: 10.1038/emboj.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy PH, Shirendeb UP. Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington’s disease. Biochim. Biophys. Acta. 2012;1822:101–110. doi: 10.1016/j.bbadis.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bates G. Huntingtin aggregation and toxicity in Huntington’s disease. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Li SH, Johnston H, Shelbourne PF, Li XJ. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat. Genet. 2000;25:385–389. doi: 10.1038/78054. [DOI] [PubMed] [Google Scholar]
- 8.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 9.Orr AL, et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu ZX, et al. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington’s disease. J. Neurosci. 2003;23:2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song W, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker MJ, Frazier AE, Gulbis JM, Ryan MT. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 2007;17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 14.MacKenzie JA, Payne RM. Mitochondrial protein import and human health and disease. Biochim. Biophys. Acta. 2007;1772:509–523. doi: 10.1016/j.bbadis.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Li SH, Yu ZX, Shelbourne P, Li XJ. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington’s disease mice. J. Neurosci. 2001;21:8473–8481. doi: 10.1523/JNEUROSCI.21-21-08473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panov AV, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 17.Rockabrand E, et al. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum. Mol. Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 18.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 19.Browne SE, et al. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann. Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 20.Gu M, et al. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann. Neurol. 1996;39:385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 21.Browne SE, Beal MF. The energetics of Huntington’s disease. Neurochem. Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 22.Ona VO, et al. Inhibition of caspase-1 slows disease progression in a mouse model of Huntington’s disease. Nature. 1999;399:263–267. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, et al. Minocycline inhibits caspase-independent and -dependent mitochondrial cell death pathways in models of Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2003;100:10483–10487. doi: 10.1073/pnas.1832501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum. Mol. Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, et al. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum. Mol. Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy PH, et al. Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer’s disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim. Biophys. Acta. 2012;1822:639–649. doi: 10.1016/j.bbadis.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costa V, et al. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol. Med. 2010;2:490–503. doi: 10.1002/emmm.201000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gines S, et al. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington’s disease knock-in mice. Hum. Mol. Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 29.Mochel F, et al. Early alterations of brain cellular energy homeostasis in Huntington disease models. J. Biol. Chem. 2012;287:1361–1370. doi: 10.1074/jbc.M111.309849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J. Clin. Invest. 2011;121:493–499. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acevedo-Torres K, et al. Mitochondrial DNA damage is a hallmark of chemically induced and the R6/2 transgenic model of Huntington’s disease. DNA Repair (Amst.) 2009;8:126–136. doi: 10.1016/j.dnarep.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirendeb UP, et al. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum. Mol. Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meisinger C, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Altmann K, Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5410–5417. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milnerwood AJ, Raymond LA. Early synaptic pathophysiology in neurodegeneration: insights from Huntington’s disease. Trends Neurosci. 2010;33:513–523. doi: 10.1016/j.tins.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Johri A, Beal MF. Antioxidants in Huntington’s disease. Biochim. Biophys. Acta. 2012;1822:664–674. doi: 10.1016/j.bbadis.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer’s disease brain is associated with mitochondrial dysfunction. J. Neurosci. 2006;26:9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q, et al. ALS-linked mutant superoxide dismutase 1 (SOD1) alters mitochondrial protein composition and decreases protein import. Proc. Natl. Acad. Sci. USA. 2010;107:21146–21151. doi: 10.1073/pnas.1014862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, et al. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 40.Roesch K, Curran SP, Tranebjaerg L, Koehler CM. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a–TIMM13 complex. Hum. Mol. Genet. 2002;11:477–486. doi: 10.1093/hmg/11.5.477. [DOI] [PubMed] [Google Scholar]
- 41.Ahting U, et al. Neurological phenotype and reduced lifespan in heterozygous Tim23 knockout mice, the first mouse model of defective mitochondrial import. Biochim. Biophys. Acta. 2009;1787:371–376. doi: 10.1016/j.bbabio.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Vögtle FN, Meisinger C. Sensing mitochondrial homeostasis: the protein import machinery takes control. Dev. Cell. 2012;23:234–236. doi: 10.1016/j.devcel.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Shao J, Diamond MI. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum. Mol. Genet. 2007;16:R115–R123. doi: 10.1093/hmg/ddm213. [DOI] [PubMed] [Google Scholar]
- 44.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 45.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 46.Weydt P, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Humphries AD, et al. Dissection of the mitochondrial import and assembly pathway for human Tom40. J. Biol. Chem. 2005;280:11535–11543. doi: 10.1074/jbc.M413816200. [DOI] [PubMed] [Google Scholar]
- 48.Trettel F, et al. Dominant phenotypes produced by the HD mutation in STHdhQ111 striatal cells. Hum. Mol. Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 49.Ehrlich ME, et al. ST14A cells have properties of a medium-size spiny neuron. Exp. Neurol. 2001;167:215–226. doi: 10.1006/exnr.2000.7551. [DOI] [PubMed] [Google Scholar]
- 50.Kristian T. Isolation of mitochondria from the CNS. Curr. Protoc. Neurosci. 2010;7:7.22. doi: 10.1002/0471142301.ns0722s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baranov SV, Stavrovskaya IG, Brown AM, Tyryshkin AM, Kristal BS. Kinetic model for Ca2+-induced permeability transition in energized liver mitochondria discriminates between inhibitor mechanisms. J. Biol. Chem. 2008;283:665–676. doi: 10.1074/jbc.M703484200. [DOI] [PubMed] [Google Scholar]
- 52.Yano M, Hoogenraad N, Terada K, Mori M. Identification and functional analysis of human Tom22 for protein import into mitochondria. Mol. Cell. Biol. 2000;20:7205–7213. doi: 10.1128/mcb.20.19.7205-7213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terada K, et al. Participation of the import receptor Tom20 in protein import into mammalian mitochondria: analyses in vitro and in cultured cells. FEBS Lett. 1997;403:309–312. doi: 10.1016/s0014-5793(97)00070-7. [DOI] [PubMed] [Google Scholar]
- 54.Kim AH, et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.