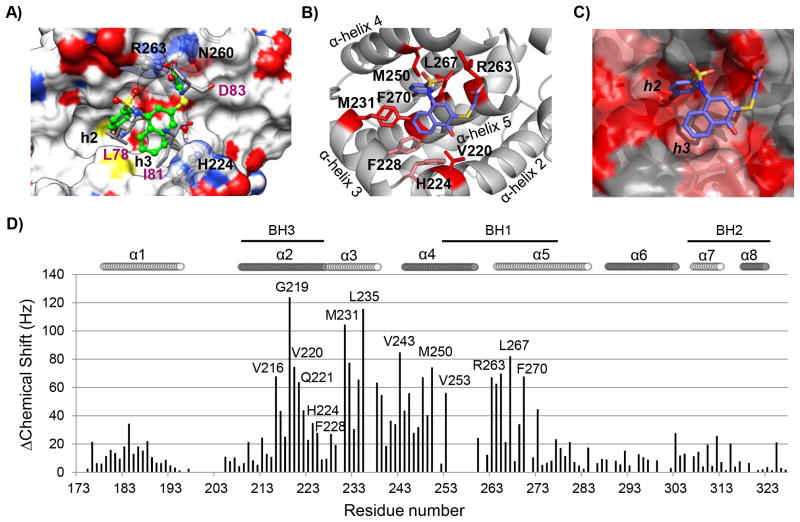
Fig. 2. Computational and structural binding studies of 2 (UMI-77) with Mcl-1.
A) Computational predicted binding pose of 2 (UMI-77) with Mcl-1 using the mNoxa BH3 peptide-bound Mcl-1 structure (PDB ID 2NLA). Superposition of the predicted binding pose of 2 (UMI-77) onto the crystal structure of mNoxa BH3 peptide with Mcl-1 (conserved residues from mNoxa, L78, I81 and D83, are presented in pink color). Hydrogen bonds with R263 and H224 from Mcl-1 protein are shown in magenta dotted lines. B) Chemical shift mapping on the predicted binding model of 2 (UMI-77) to Mcl-1. The side chains of the residues involved in the interactions with UMI-77 and confirmed with the HSQC NMR studies are shown and labeled. C) Chemical shift mapping using a surface representation of the predicted binding model between UMI-77 and Mcl-1 (chemical shifts ≥60 Hz are labeled in red, shifts between 30 and 60 Hz are labeled in pink). D) Chemical shift differences for Mcl-1 in presence of 2 (UMI-77) (2 equivalents) against residue number.