Abstract
The plant pathogenic bacterial genus Pectobacteirum consists of heterogeneous strains. The P. carotovorum species is a complex strain showing divergent characteristics, and a new subspecies named P. carotovorum subsp. brasiliensis has been identified recently. In this paper, we re-identified the P. carotovorum subsp. brasiliensis isolates from those classified under the subspecies carotovorum and newly isolated P. carotovorum subsp. brasiliensis strains. All isolates were able to produce plant cell-wall degrading enzymes such as pectate lyase, polygalacturonase, cellulase and protease. We used genetic and biochemical methods to examine the diversity of P. carotovorum subsp. brasiliensis isolates, and found genetic diversity within the brasiliensis subsp. isolates in Korea. The restriction fragment length polymorphism analysis based on the recA gene revealed a unique pattern for the brasiliensis subspecies. The Korean brasiliensis subsp. isolates were divided into four clades based on pulsed-field gel electrophoresis. However, correlations between clades and isolated hosts or year could not be found, suggesting that diverse brasiliensis subsp. isolates existed.
Keywords: 16S rDNA, Pectobacterium catorovorum subsp. brasiliensis, PFGE, recA, RFLP
Bacteria belonging to Pectobacterium carotovorum cause soft rot disease in diverse vegetables and crops worldwide. These bacteria produce several different plant cell-wall degrading enzymes (PCWDEs) such as pectinase, polygalacturonase (Peh) and cellulase (Cel). The maceration of cell walls caused by PCWDEs results in soft rot symptoms in host plants. Since Pectobacterium species can produce PCWDEs, soft rot disease can occur in the field as well as during transportation and storage. P. carotovorum subsp. carotovorum has a wide host range including potato, carrot, lettuce, onion and Chinese cabbage, but P. atrosepticum, P. betavasculorum, P. wasabiae and P. carotovorum subsp. odoriferum have a relatively narrow host range that includes potato, sugarbeet, horseradish and chicory, respectively (Charkowski et al., 2006; Seo et al., 2005; Toth et al., 2003).
Pectobacterium is a complex taxon of strains with diverse characteristics. Recently, based on new molecular biology techniques, several species in the former Erwinia genus have been re-classified as Erwinia, Brenneria, Pectobacterium, Dickeya and Pantoea (Samson et al., 2005; Waleron et al., 2002). As a result, Erwinia carotovora was renamed Pectobacterium carotovorum. In recent years, P. carotovorum species was classified into five subspecies, atrosepticum, betavasculorum, carotovorum, odoriferum and wasabiae (Hauben et al., 1998). However, the genetic differences among the subspecies atrosepticum, betavasculroum and wasabiae were too significant to be placed within the same species P. carotovorum. Gardan et al. (2003) suggested the three subpecies atrosepticum, betavasculorum and wasabiae should be elevated to the species level and not subspecies.
Recently, bacteria isolated from the blackleg-diseased potato in Brazil were similar to P. carotovorum subsp. carotovorum that usually cause soft rot disease, but not similar to P. atrosepticum, the major pathogen of the blackleg disease of potato (Duarte et al., 2004). The bacteria were termed P. carotovorum subsp. brasiliensis, a new subspecies of P. carotovorum (Duarte et al., 2004; Nahan et al., 2012). After this first report of the subspecies brasiliensis, scientists in several countries, such as the USA, South Africa, Canada, New Zealand and Korea, have reported its existence (Choi and Kim, 2013; De Boer et al., 2012; Kim et al., 2009; Panda et al., 2012; van der Merwe et al., 2010).
The first isolation of P. carotovorum subsp. brasiliensis in Korea was reported in 2013 (Choi and Kim, 2013). In 1999, 11 bacterial strains isolated from blackleg diseased potato reportedly showed different characteristics from P. carotovorum subsp. carotovorum or P. atrosepticum strains (Seo et al., 2004, 2005). However, the study did not investigate whether that strain had been existed in Korea or not. Therefore, this research was conducted to demonstrate that the subspecies brasiliensis existed previously in Korea, and was not a new subspecies imported recently.
Materials and Methods
Bacterial strains, media and cultivation conditions
P. carotovorum subsp. brasiliensis strains used in this study are listed in Table 1. The strains were isolated from various hosts showing soft rot disease symptoms between 1997 and 2013 in Korea. The strains were stored at −80°C until use. For experiments, the strains were cultured on tryptic soy agar (TSA, BD, Franklin Lakes, NJ, USA) or in tryptic soy broth (TSB, BD) medium at 28°C, and then applied to tests.
Table 1.
List of Pectobacterium carotovorum strains and 16S rDNA sequence analysis results
| Previous strain name | Strain no. | Year isolated | Isolated from | 16S rDNA analysis (assigned name) |
|---|---|---|---|---|
| P. carotovorum subsp. | P. carotovorum subsp. | |||
| carotovotum | 30 | 1997 | Chinese cabbage | brasiliensis |
| carotovotum | 34 | 1997 | Chinese cabbage | brasiliensis |
| carotovotum | 35 | 1997 | Chinese cabbage | brasiliensis |
| carotovotum | 36 | 1997 | Chinese cabbage | brasiliensis |
| carotovotum | 49 | 1997 | Chinese cabbage | brasiliensis |
| carotovotum | E3 | 1997 | Tomato | brasiliensis |
| carotovotum | E10 | 1997 | Potato | brasiliensis |
| carotovotum | E35 | 1997 | Chinese cabbage | brasiliensis |
| carotovotum | 52 | 1997 | Chinese cabbage | brasiliensis |
| carotovotum | E12 | 1997 | Chinese cabbage | brasiliensis |
| carotovotum | 76 | 1998 | Chinese cabbage | brasiliensis |
| carotovotum | 113 | 2000 | Chinese cabbage | brasiliensis |
| carotovotum | 123 | 2000 | Chinese cabbage | brasiliensis |
| carotovotum | 112 | 2000 | Chinese cabbage | brasiliensis |
| carotovotum | E42 | 2010 | Chrysanthemum | brasiliensis |
| carotovotum | E52 | 2012 | Eggplant | brasiliensis |
| carotovotum | E53 | 2012 | Eggplant | brasiliensis |
| brasiliensis | N1 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N2 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N3 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N4 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N5 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N6 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N7 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N8 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N9 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N10 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N11 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N12 | 2013 | Nepenthes | brasiliensis |
| brasiliensis | N13 | 2013 | Nepenthes | brasiliensis |
Pathogenicity test
The pathogenicity test against Chinese cabbage was performed according to a previously published procedure with some modification (Lee et al., 2013a). The fresh strains grown on TSA medium for 24–48 h were harvested by a sterilized cotton swab and resuspended in dilution solution (10 mM MgCl2) to an approximate cell density of 108 cfu/mL. Ten microliters of each suspension was inoculated into Chinese cabbage, and then soft rot symptoms were observed while the Chinese cabbage was incubated at 28°C in sterilized plastic boxes with sufficient moisture. The Chinese cabbage inoculated with dilution solution was used as a negative control.
Identification of bacteria using 16S rDNA sequence analysis
After growth of the strains on TSA medium, 16S rRNA gene was sequenced by Macrogen, Inc. (Korea). The strains were identified based on a comparison of their nucleotide sequences with those in the GenBank database using the BLAST software on the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov/).
PCWDE assays
Extracellular enzyme assays were performed according to procedures described previously (Lee et al., 2013b). To prepare samples for the enzyme assays, the strains were cultured overnight in TSB medium at 28°C and cells were removed by centrifugation at 15,000 × g for 10 min. Supernatants were poured into wells on each assay plate. The following substances were used for semiquantitative enzyme assay media: 1% polygalacturonic acid (PGA) for pectate lyase (Pel) and Peh; 1% carboxymethyl cellulose for Cel; 1% skim milk for protease (Prt) assay. Pel and Peh assay media were developed with 4 N HCl and Prt media with 0.1% Congo Red.
recA PCR-restriction fragment length polymorphism (RFLP) analysis
Genomic DNA of the strains was extracted using the G-spin genomic DNA isolation kit (iN-tRON Biotechnology, Seongnam, Korea), and then stored at −20°C until needed. The recA gene was amplified with the recA-specific primers Ec_recA_F (GGTAAAGGGTCTATCATGCG) and Ec_recA_R (CCTTCACCATACATAATTTGGA) (Waleron et al., 2002). The ∼730 bp amplified products were digested with the restriction enzymes TasI, Tru1I, AluI and HinfI (Takara or Fermentas). The digested fragments were separated in a 2.0% agarose gel in Tris-acetate-EDTA buffer and visualized for further band pattern analysis.
Pulsed-field gel electrophoresis (PFGE) analysis
In accordance with the standardized PulseNet protocol maintained at the Center for Disease Control and Prevention, we performed pulsed-field gel electrophoresis (Hunter et al., 2005). Strains grown on TSA medium were collected by a cotton swab and then resuspended in 1 ml of Tris-EDTA buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) to an OD600nm of 0.5–0.7. After lysozyme and proteinase K treatments, plugs were incubated in EC lysis buffer (6 mM Tris–HCl, 1 M NaCl, 100 mM EDTA, 0.5% Brij-58, 0.2% sodium deoxycholate, 0.5% sodium lauroly sarcosine). The plugs were digested with SmaI (Takara), and the genomic DNA fragments were separated in a 1% Seakem gold agarose gel using a CHEF Mapper (Bio-Rad, Hercules, CA, USA) system. Gels were stained with SYBR® Gold Stain (Invitrogen, USA). The cellular DNA from Salmonella serotype Braenerup H9812 was treated with XbaI (Takara) and that DNA fragments were used as a standard for normalization (Hunter et al., 2005). Using the Fingerprinting II software (Bio-Rad), PFGE band patterns were analyzed, and a dendrogram was clustered and analyzed using the un-weighted pair group method of average linkage (UPGMA).
Identification of strains using the Biolog system
The carbon source usage of the isolated strains was determined using a microstation system with a GN2 MicroPlate (Biolog, Hayward, CA, USA) according to a procedure published previously (Holmes et al., 1994). Briefly, overnight cultures of bacteria on TSA plates were resuspended in 0.85% saline solution and 150 μl of suspension were dispensed per microplate well. The results obtained using the Biolog microstation set were compared to a given value provided by the Microlog software (Biolog).
Indole production test
The indole spot test was used to determine indole production. Strains streaked on TSA medium were incubated for 18–24 h at 28°C. A Whatman filter paper was wetted with Kovács reagent (10 g p-dimethylaminobenzaldehyde, 50 ml HCl and 150 ml isoamyl alcohol). After smearing the strains on the filter paper, the development of a red/pink color was observed.
Results and Discussion
Identification of strains belonging to P. carotovorum subsp. brasiliensis.
Because a new subspecies brasiliensis of the P. carotovorum species was reported in Korea in 2013 (Choi and Kim, 2013), determining whether this new subspecies was imported or existed previously in Korea is important. A total of 128 isolates, identified as P. carotovorum subsp. carotovorum collected in 1997, 1998, 1999 and 2000 were maintained at −80°C, reisolated and reidentified. Additionally, eight strains were isolated from various hosts including chrysanthemum and eggplant in 2010 and 2012, and 13 strains from nepenthes in 2013 were used for identification of the subspecies brasiliensis. All strains used in this present study were tested for their pathogenicity against Chinese cabbage to determine whether the preserved strains maintained their characteristics. As expected, all tested strains caused severe soft rot disease against the host (data not shown).
Because sequence analysis of 16S rRNA genes of bacterial strains was successfully used to classify bacterial isolates in P. carotovorum species into the subspecies level (Hauben et al., 1998), the 16S rDNA sequences of all 149 strains were identified and analyzed with the NCBI Blast program. A total of 30 isolates analyzed were identified as subspecies brasiliensis with 99% identity at the nucleotide level (Table 1). Among 30 isolates, 10 were isolated in 1997, 1 in 1998, 3 in 2000, and 1 in 2010. Because the first report of the subspecies brasiliensis was in 1999 (Duarte et al., 2004) and Korean isolates reported by Choi and Kim (2013) were collected in 2011, these data suggest that P. carotovorum subsp. brasiliensis is not a newly imported subspecies, but existed previously in Korea as P. carotovorum subsp. carotovorum and could not be identified due to a lack of phylogenetic information.
The PCWDE production of the subspecies brasiliensis isolates
Since bacteria of the genus Pectobacterium produce and secrete PCWDEs, macerate plant cell walls and then cause soft rot disease, the PCWDEs is an important virulence factor of the plant pathogenic bacteria. The P. carotovorum subsp. carotovorum Pcc21 strain has numerous genes involved in the production of several PCWDEs; mutants lacking enzyme production or secretion ability are also non-pathogenic (Lee et al., 2013b; Park et al., 2012). PCWDE production is also related to the plant defense response (Lee et al., 2008; Mäe et al., 2001).
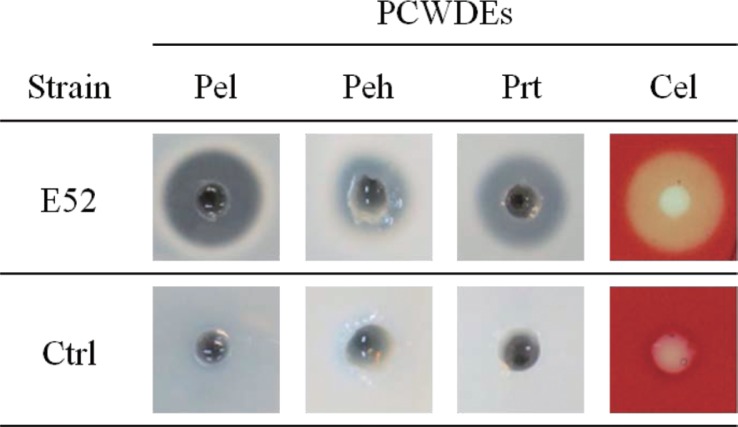
Since the production and secretion of PCWDEs is an important characteristic of the genus Pectobacterium, the production and secretion of four PCWDEs in the strains were surveyed. All tested strains of the subspecies brasiliensis could produce and secrete the four PCWDE: Pel, Peh, Cel, and Prt (Table 2 and Fig. 1). The production and secretion of the major PCWDEs and the pathogenicity to Chinese cabbage suggested that all of the newly identified strains belonged to the subspecies brasiliensis and had strong pathogenicity in vegetables, in which they caused soft rot disease.
Table 2.
Plant cell-wall degrading enzyme (PCWDE) activities of P. carotovorum subsp. brasiliensis
| Strain namea | Strain no. | PCWDEsb
|
Indole production | |||
|---|---|---|---|---|---|---|
| Pel | Peh | Cel | Prt | |||
| P. carotovorum subsp. | ||||||
| brasiliensis | 30 | + | + | + | + | + |
| brasiliensis | 34 | + | + | + | + | + |
| brasiliensis | 35 | + | + | + | + | + |
| brasiliensis | 36 | + | + | + | + | + |
| brasiliensis | 49 | + | + | + | + | + |
| brasiliensis | E3 | + | + | + | + | + |
| brasiliensis | E10 | + | + | + | + | + |
| brasiliensis | E35 | + | + | + | + | + |
| brasiliensis | 52 | + | + | + | + | + |
| brasiliensis | E12 | + | + | + | + | + |
| brasiliensis | 76 | + | + | + | + | + |
| brasiliensis | 113 | + | + | + | + | + |
| brasiliensis | 123 | + | + | + | + | + |
| brasiliensis | 112 | + | + | + | + | + |
| brasiliensis | E42 | + | + | + | + | + |
| brasiliensis | E52 | + | + | + | + | + |
| brasiliensis | E53 | + | + | + | + | + |
| brasiliensis | N1 | + | + | + | + | + |
| brasiliensis | N2 | + | + | + | + | + |
| brasiliensis | N3 | + | + | + | + | + |
| brasiliensis | N4 | + | + | + | + | + |
| brasiliensis | N5 | + | + | + | + | + |
| brasiliensis | N6 | + | + | + | + | + |
| brasiliensis | N7 | + | + | + | + | + |
| brasiliensis | N8 | + | + | + | + | + |
| brasiliensis | N9 | + | + | + | + | + |
| brasiliensis | N10 | + | + | + | + | + |
| brasiliensis | N11 | + | + | + | + | + |
| brasiliensis | N12 | + | + | + | + | + |
| brasiliensis | N13 | + | + | + | + | + |
Newly assigned strain name in Table 1 was used.
Pel, pectate lyase; Peh, polygalaturonase; Cel, cellulase; Prt, protease.
Fig. 1.
Plant cell-wall degrading enzyme (PCWDE) activities of P. carotovorum subsp. brasiliensis strains isolated in Korea. The supernatant of P. carotovorum subsp. brasiliensis strain culture was transferred to pectate lyase (Pel), polygalacturonase (Peh), protease (Prt), and cellulase (Cel) assay media. The photographs are representative of three independent experiments using P. carotovorum subsp. brasiliensis E52. Other strains used in this study showed similar positive activities. Tryptic soy broth was used as the negative control (Ctrl); no enzyme activity was observed.
Classification of the subspecies brasiliensis isolates by PCR-RFLP
A total of 30 isolates were identified as subspecies brasiliensis and 11 were isolated before the first subspecies brasiliensis isolates were reported. To investigate the genetic diversity of the subspecies brasiliensis isolates in Korea, the recA gene restriction enzyme digestion patterns of all 30 bacteria were analyzed. The sequence analysis of 16S and 23S rRNA, rpoS, gyrA, recA and groEL genes has been used to classify plant pathogenic bacteria (Waleron et al., 2002). Additionally, in bacteria of the genus Pectobacterium, the pel gene has been used as a marker (Darrasse et al., 1994). The recA gene encodes the RecA protein, which is involved in the DNA repair system and SOS response; the recA gene DNA sequence is well conserved (Eisen et al., 1995). The RFLP patterns of the recA gene of the genus Pectobacterium using several restriction enzymes are available (Waleron et al., 2002; 2008).
Based on previous reports, the PCR-amplified recA DNA fragment was digested using four restriction enzymes (TasI, Tru1I, AluI and HinfI) separately and the fragment patterns were analyzed. The Korean brasiliensis isolates were not highly genetically divergent (Table 3). The RFLP patterns of the 30 different subspecies brasiliensis isolates were divided into group 8 and group D. Based on the Waleron’s pattern designation (Waleron et al., 2002), RFLP group 8 showed the typical pattern for subspecies carotovorum and group D exhibited a new pattern for the Korean subspecies brasiliensis isolates. Although the Korean isolates were divided into two groups, group D showed a different pattern with only one of the restriction enzymes used. The genetic variation among Korean subspecies brasiliensis isolates was minimal, suggesting that differentiation may have begun only recently. However, further in-depth investigations should be conducted to determine whether group D is a pattern unique to the Korean strains and if group 8 should be regarded as a pattern of the subspecies brasiliensis and not the subspecies carotovorum.
Table 3.
Restriction fragment length polymorphism (RFLP) analysis of P. carotovorum subsp. brasiliensis based on the recA gene
| Strain namea | Strain no. | Year isolated | Isolated from |
recA RFLP patternb
|
RFLP groupc | |||
|---|---|---|---|---|---|---|---|---|
| TasI | Tru1I | AluI | HinfI | |||||
| P. carotovorum subsp. | ||||||||
| brasiliensis | 30 | 1997 | Chinese cabbage | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | 34 | 1997 | Chinese cabbage | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | 35 | 1997 | Chinese cabbage | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | 36 | 1997 | Chinese cabbage | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | 49 | 1997 | Chinese cabbage | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | E3 | 1997 | Tomato | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | E10 | 1997 | Potato | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | E35 | 1997 | Chinese cabbage | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | 52 | 1997 | Chinese cabbage | 6 | 3 | 2 | 2 | D |
| brasiliensis | E12 | 1997 | Chinese cabbage | 6 | 3 | 2 | 2 | D |
| brasiliensis | 76 | 1998 | Chinese cabbage | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | 113 | 2000 | Chinese cabbage | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | 123 | 2000 | Chinese cabbage | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | 112 | 2000 | Chinese cabbage | 6 | 3 | 2 | 2 | D |
| brasiliensis | E42 | 2010 | Chrysanthemum | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | E52 | 2012 | Eggplant | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | E53 | 2012 | Eggplant | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N1 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N2 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N3 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N4 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N5 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N6 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N7 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N8 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N9 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N10 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N11 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N12 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
| brasiliensis | N13 | 2013 | Nepenthes | 4 | 3 | 2 | 2 | 8 |
Newly assigned strain name in Table 1 was used.
PCR-RFLP pattern and group number were acquired from the previous report (Waleron et al., 2002). RFLP group D was newly designated in this research.
Classification of the subspecies brasiliensis isolates using PFGE
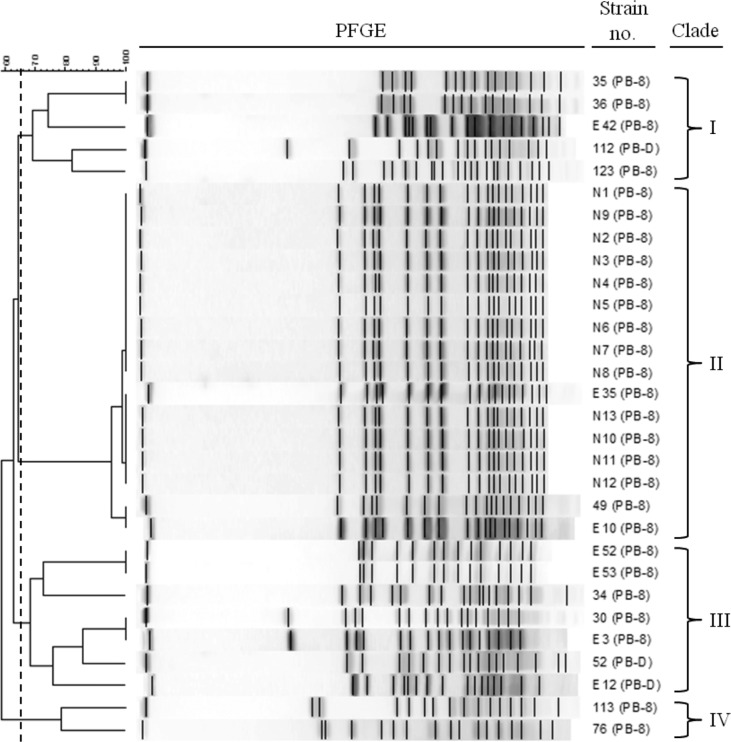
Because the analysis of RFLP based on the recA gene of the Korean subspecies brasiliensis isolates showed two different patterns, further investigations of the genetic diversity of the isolates were needed. Since the recA-based RFLP used a small DNA fragment (∼730 bp), PFGE was used to investigate the diversity of Korean subspecies brasiliensis isolates and analyze the entire genome. PFGE was used for the identification of bacterial subspecies, because this method was highly sensitive and showed stable genetic differentiation of isolated bacteria (Hunter et al., 2005). The PFGE results after SmaI restriction enzyme digestion of the total DNA from each subspecies brasiliensis isolate showed more diverse genetic variation (Fig. 2). Analysis of the PFGE results using the Fingerprinting II software (Bio-Rad) indicated that the 30 Korean subspecies brasiliensis isolates grouped into four clades. However, no correlations between the clade and host or year of isolation were found. Therefore, diverse subspecies brasiliensis isolates existed previously in Korea, and new developments in technology facilitated their differentiation from the subspecies carotovorum.
Fig. 2.
Dendrogram of the pulsed-field gel electrophoresis (PFGE) results of P. carotovorum subsp. brasiliensis strains isolated in Korea. Four PFGE clades (I–IV) were detected based on a similarity cut-off of 65% (0.5% optimization and 1.0% tolerance). Note that PB is P. carotovorum subsp. brasiliensis, and 8 and D are recA-based RFLP groups in Table 3.
Biochemical characteristics of the Korean subspecies brasiliensis isolates
Assays of carbon source utilization using the Biolog system and indole production of Korean subspecies brasiliensis isolates were conducted. Based on the results, the Biolog software identified the subspecies brasiliensis isolates as subspecies carotovorum (data not shown). In a previous report, subspecies brasiliensis strains were included in the heterogeneous and broader subspecies carotovorum group (Duarte et al., 2004). These results confirmed that carbon source utilization analysis is not sufficient to distinguish subspecies brasiliensis isolates from those of subspecies carotovorum, although several differences in biological characteristics exist. Therefore, distinguishing subspecies brasiliensis from the subspecies carotovorum is difficult.
The ability of certain bacteria to form indole from the amino acid tryptophan is an important phenotypic characteristic. Demonstration of indole production is a useful characterization test. All tested Korean subspecies brasiliensis isolates could produce indole (Table 2). This result is inconsistent with a previous report that isolates of the sub-species brasiliensis are negative for indole production (Choi and Kim, 2013; Pasanen et al., 2013). In recent studies, subspecies brasiliensis strains were divided into two subgroups using phylogenetic analysis based on multilocus sequence analysis of eight housekeeping gene fragments, and showed diverse phenotypic characteristics (Nabhan et al., 2012); one third of the subspecies carotovorum isolates could form indole (Seo et al., 2002; Yap et al., 2004). Thus, these results suggest that not all Korean subspecies brasiliensis isolates can form indole; therefore, variants exist among isolates of the Korean subspecies brasiliensis.
In this present study, we reclassified several isolates as P. carotovorum subsp. brasiliensis previously identified as P. carotovorum subsp. carotovorum, and isolated subspecies brasiliensis strains from various plants. Genetic analyses and biochemical tests indicated the genetic diversity of P. carotovorum subsp. brasiliensis strains isolated in Korea. Based on our results, we suggest that the newly identified P. carotovorum subsp. brasiliensis was not imported from a foreign country and previously existed in Korea as much as 20 years ago; further intensive comparison studies should be conducted. Knowledge of the genetic differentiation of the subspecies carotovorum and brasiliensis is important. The reported results will facilitate determination of the host specificity of plant pathogens.
Acknowledgments
This work was supported by the Rural Development Administration (RDA) fund PJ008639. D. H. L. and J.–A L. were supported by the postdoctoral fellowship program of the National Academy of Agricultural Science (NAAS), RDA, Republic of Korea.
References
- Charkowski AO. The soft rot Erwinia. In: Gnanamanickam SS, editor. Plant-Associated Bacteria. Springer; Netherlands: 2006. pp. 423–505. [Google Scholar]
- Choi O, Kim J. Pectobacterium carotovorum subsp. brasiliense causing soft rot on paprika in Korea. J Phytopathol. 2013;161:125–127. [Google Scholar]
- Darrasse A, Priou S, Kotoujansky A, Bertheau Y. PCR and restriction length polymorphism of a pel gene as a tool to identify Erwinia carotovora in relation to potato diseases. Appl Environ Microbiol. 1994;60:1437–1443. doi: 10.1128/aem.60.5.1437-1443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer SH, Li X, Ward LJ. Pectobacterium spp. associated with bacterial stem rot syndrome of potato in Canada. Phytopathology. 2012;102:937–947. doi: 10.1094/PHYTO-04-12-0083-R. [DOI] [PubMed] [Google Scholar]
- Duarte V, De Boer SH, Ward LJ, De Oliveira AMR. Characterization of atypical Erwinia carotovora strains causing blackleg of potato in Brazil. J Appl Microbiol. 2004;96:535–545. doi: 10.1111/j.1365-2672.2004.02173.x. [DOI] [PubMed] [Google Scholar]
- Eisen JA. The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardan L, Gouy C, Christen R, Samson R. Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Int J Syst Evol Microbiol. 2003;53:381–391. doi: 10.1099/ijs.0.02423-0. [DOI] [PubMed] [Google Scholar]
- Hauben L, Moore ERB, Vauterin L, Steenackers M, Mergaert J, Verdonck L, Swings J. Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst Appl Microbiol. 1998;21:384–397. doi: 10.1016/S0723-2020(98)80048-9. [DOI] [PubMed] [Google Scholar]
- Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005;43:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-S, Ma B, Perna NT, Charkowski AO. Phylogeny and virulence of naturally occurring type III secretion system-deficient Pectobacterium strains. Appl Environ Microbiol. 2009;75:4539–4549. doi: 10.1128/AEM.01336-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Kim J-B, Kim M, Roh E, Jung K, Choi M, Oh C, Choi J, Yun J, Heu S. Microbiota on spoiled vegetables and their characterization. J Food Prot. 2013a;76:1350–1358. doi: 10.4315/0362-028X.JFP-12-439. [DOI] [PubMed] [Google Scholar]
- Lee DH, Lim J-A, Lee J, Roh E, Jung K, Choi M, Oh C, Ryu S, Yun J, Heu S. Characterization of genes required for the pathogenicity of Pectobacterium carotovorum subsp. carotovorum Pcc21 in Chinese cabbage. Microbiology. 2013b;159:1487–1496. doi: 10.1099/mic.0.067280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Hong JB, Hong SB, Choi M-S, Jeong KY, Park H-J, Hwang D-J, Lee S, Ra D, Heu S. Disease-resistant transgenic Arabidopsis carrying the expI gene from Pectobacterium carotovorum subsp. carotovorum SL940. Plant Pathol J. 2008;24:183–190. [Google Scholar]
- Mäe A, Montesano M, Koiv V, Palva ET. Transgenic plants producing the bacterial pheromone N-acylhomoserine lactone exhibit enhanced resistance to the bacterial phytopathogen Erwinia carotovora. Mol Plant-Microbe Interact. 2001;14:1035–1042. doi: 10.1094/MPMI.2001.14.9.1035. [DOI] [PubMed] [Google Scholar]
- Nabhan S, De Boer SH, Maiss E, Wydra K. Taxonomic relatedness between Pectobacterium carotovorum subsp. carotovorum, Pectobacterium carotovorum subsp. odoriferum and Pectobacterium carotovorum subsp. brasiliense subsp. nov. J Appl Microbiol. 2012;113:904–913. doi: 10.1111/j.1365-2672.2012.05383.x. [DOI] [PubMed] [Google Scholar]
- Panda P, Fiers MAWJ, Armstrong K, Pitman AR. First report of blackleg and soft rot of potato caused by Pectobacterium carotovorum subsp. brasiliensis in New Zealand. New Dis Rep. 2012;26:15. [Google Scholar]
- Park T-H, Choi B-S, Choi A-Y, Choi I-Y, Heu S, Park B-S. Genome sequence of Pectobacterium carotovorum subsp. carotovorum strain PCC21, a pathogen causing soft rot in Chinese cabbage. J Bacteriol. 2012;194:6345–6346. doi: 10.1128/JB.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasanen M, Laurila J, Brader G, Palva ET, Ahola V, van der Wolf J, Hannukkala A, Pirhonen M. Characterisation of Pectobacterium wasabiae and Pectobacterium carotovorum subsp. carotovorum isolates from diseased potato plants in Finland. Ann Appl Biol. 2013;163:403–419. [Google Scholar]
- Samson R, Legendre JB, Christen R, Fischer-Le Saux M, Achouak W, Gardan L. Transfer of Pectobacterium chrysanthemi(Burkholder et al., 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int J Syst Evol Microbiol. 2005;55:1415–1427. doi: 10.1099/ijs.0.02791-0. [DOI] [PubMed] [Google Scholar]
- Seo ST, Koo JH, Hur JH, Lim CK. Characterization of Korean Erwinia carotovora strains from potato and Chinese cabbage. Plant Pathol J. 2004;20:283–288. [Google Scholar]
- Seo ST, Furuya N, Lim CK, Takanami Y, Tsuchiya K. Phenotypic and genetic diversity of Erwinia carotovora ssp. carotovora strains from Asia. J Phytopathol. 2002;150:120–127. [Google Scholar]
- Seo S-T, Lee S, Lee J-S, Han K-S, Jang H-I, Lim C-K. Genetic characterization of potato blackleg strains from Jeju island. Res Plant Dis. 2005;11:140–145. [Google Scholar]
- Toth IK, Bell KS, Holeva MC, Birch PRJ. Soft rot erwiniae: from genes to genomes. Mol Plant Pathol. 2003;4:17–30. doi: 10.1046/j.1364-3703.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- van der Merwe JJ, Coutinho TA, Korsten L, van der Waals JE. Pectobacterium carotovorum subsp. brasiliensis causing blackleg on potatoes in South Africa. Eur J Plant Pathol. 2010;126:175–185. [Google Scholar]
- Waleron M, Waleron K, Podhajska AJ, Lojkowska E. Genotyping of bacteria belonging to the former Erwinia genus by PCR–RFLP analysis of a recA gene fragment. Microbiology. 2002;148:583–595. doi: 10.1099/00221287-148-2-583. [DOI] [PubMed] [Google Scholar]
- Waleron M, Waleron K, Geider K, Lojkowska E. Application of RFLP analysis of recA, gyrA and rpoS gene fragments for rapid differentiation of Erwinia amylovora from Erwinia strains isolated in Korea and Japan. Eur J Plant Pathol. 2008;121:161–172. [Google Scholar]
- Yap M-N, Barak JD, Charkowski AO. Genomic diversity of Erwinia carotovora subsp. carotovora and its correlation with virulence. Appl Environ Microbiol. 2004;70:3013–3023. doi: 10.1128/AEM.70.5.3013-3023.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]