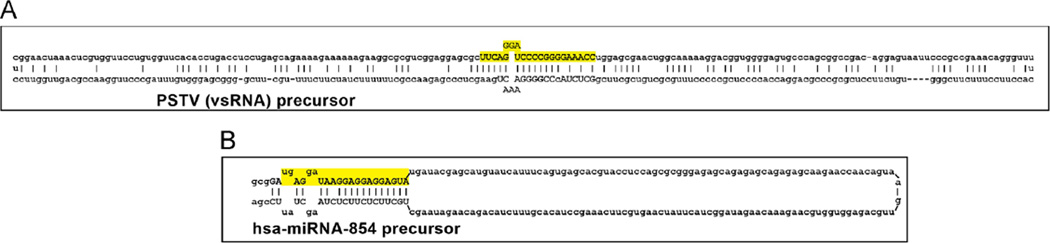
Fig. 1.
This highly schematicized figure shows the predicted structures of (A) the 359 nucleotide (nt) circular potato spindle tuber viroid (PSTV) viroid specific (vsRNA) precursor; and (B) a 214 nt hairpin-shaped precursor to the 21 nt microRNA-854 (hsa-miRNA-854; ultimately processed from the 214 nt precursor sequence and overlaid in yellow); the hsa-miRNA-854 precursor contains a large (162 nt) terminal loop structure; for both PSTV and miRNA-854 other internal free energy bonding schemes and/or secondary or tertiary structures may exist; hsa-miRNA-854 is amongst one of the most evolutionary ancient miRNAs (see text; Arteaga-Vazquez et al., 2006; Krol and Krzyzosiak, 2006; Ritchie et al., 2007; Ding, 2009; Triboulet and Gregory, 2010; unpublished). In both cases these ssRNA precursors are further processed by an RNase III of the family of Dicer-like proteins to generate smaller ssRNA species (mature vsRNA or miRNA sequences highlighted in yellow); nucleotides thought to be critical in the mature PSTV or miRNA-854 ~21–22 nt sequences are in upper case; mature vsRNAs or miRNAs have potential to alter the normal gene expression patterns in the host plant by viroids, or of mRNAs via miRNA–mRNA interactions in many plant and animal species including humans (Krol and Krzyzosiak, 2006; Arteaga-Vazquez et al., 2006; Ritchie et al., 2007; Ding, 2009; Navarro et al., 2012; Hammann and Steger, 2012). While naked ssRNAs such as these depicted have relatively short half-lives in vitro, and retinal and human brain neuronal miRNAs appear to have very limited stabilities (Sethi and Lukiw, 2009; Krol et al., 2010), viroid and miRNA half-lives may be greatly extended by single- or double-stranded RNA-binding proteins, by complex secondary and tertiary structures, by RNA circularization, by storage in vesicles, or by combinations of these and other factors (Chen and Shyu, 1995; Cui et al., 2005; Sethi and Lukiw, 2009; Krol et al., 2010; see text). Interestingly, PSTV is the smallest known self-replicating pathogen of all species; it is noteworthy that both the plant and animal kingdoms have adopted similar ‘minimalistic’ ssRNA strategies to convey highly specific- and selective-genetic regulatory information in the propagation of both homeostatic and potentially pathogenic RNA signals. Other secondary and tertiary structures are possible; PSTV and miRNA-854 ssRNA sequences and/or precursor structures were derived from GenBank accession M36163.1; GI:333356 (http://www.ncbi.nlm.nih.gov/nuccore/M36163.1) or miRBase Accession MI0005412 (http://www.mirbase.org/cgi-bin/mirnaentry.pl?acc=MI0005412); secondary structures were predicted using several freely available online web servers (such as http://mfold.rna.albany.edu/?q=mfold).