Figure 4.
Identification of Ubiquitination Machinery
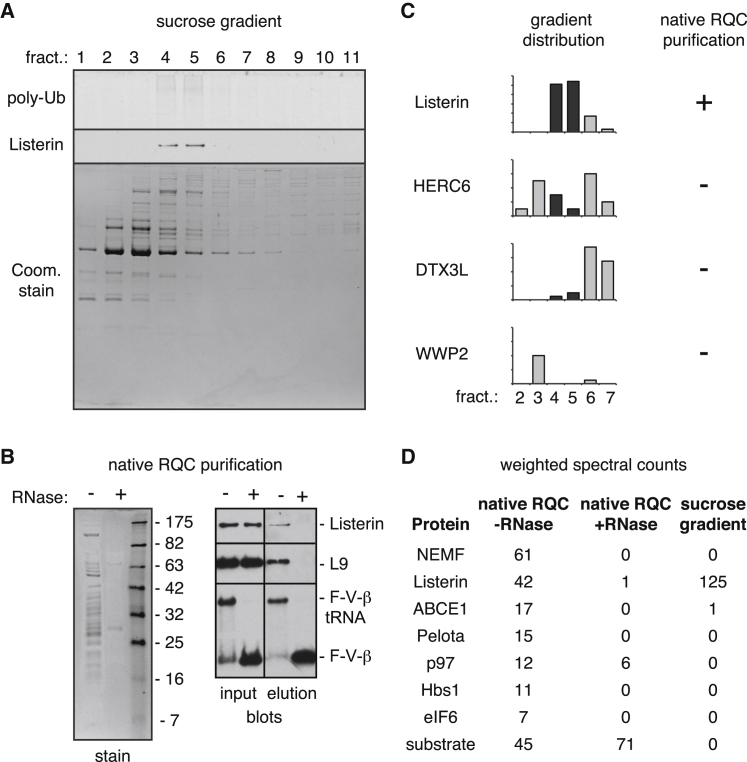
(A) Fractions from the final sucrose gradient step of a purification scheme (see Figure S3A) were analyzed for RNC ubiquitination activity (top), Listerin immunoblot (middle), and Coomassie stain (bottom).
(B) Affinity purification of in vitro translated F-VHP-β RNCs via the scheme in Figure S3B. The products purified in the absence or presence of RNase were analyzed by Coomassie staining (left) and immunoblotting (right).
(C) Abundance profiles of E3 ubiquitin ligases across gradient fractions 2–7 from (A) identified by mass spectrometry. Fractions 4 and 5, where peak RNC ubiquitination activity was observed in (A), are highlighted in black.
(D) Weighted spectral counts of select proteins identified from mass spectrometry analysis of the native RQC purification from panel B (see Table S1 for the complete list). The spectral counts for these same proteins from the sucrose gradient fractions of (A) are also shown. (See also Figure S3 and Table S1.)