Figure 3.
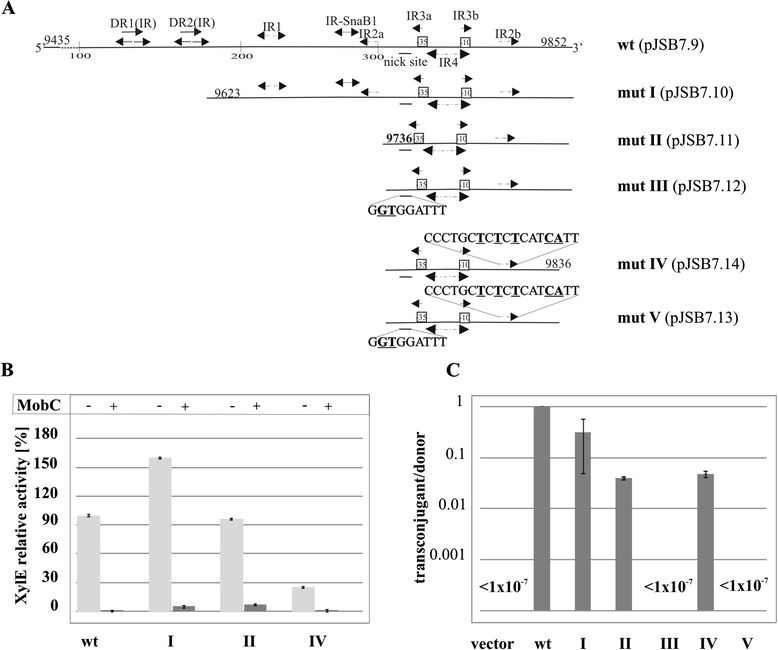
Deletion mapping of MobC binding site in the mobCp region. A. Schematic representation of deletion and point mutation derivatives. Coordinates of RA3 fragments used in the analysis and structural motifs identified in the region are shown. Sequences of modified motifs (nick site and IR2b) are shown with the modifications underlined and in bold. B. MobC regulation of mobCp in vivo. DH5α strains carrying pJSB7.9 (mobC p- xylE) or mobCp mutant derivatives (labeled according to scheme A) were transformed with empty expression vector pGBT30 or with pJSB5.1 tacp-mobC. Diagram presents XylE activities assayed in extracts of the various double transformants relative to the XylE activity detected in extract of control strain DH5α(pJSB7.9)(pGBT30). Light grey bars correspond to the results obtained for the double transformants with derivatives of pPT01 and the empty vector pGBT30, dark grey bars demonstrate results obtained for thestrains with the same pPT01 derivatives but with pJSB5.1 tacp-mobC. Mean values with standard deviation of at least three assays are shown. C. DH5α(RA3) strain was transformed with pPT01 derivatives carrying deletion variants of mobCp region. Double transformants were used as donors in conjugation with DH5α RifR strain as the recipient. The frequency of mobilization is indicated on a semi-logarithmic scale as the number of transconjugants/donor cells, where vector corresponds to empty pPT01, wt to pJSB7.9, and roman numerals to the mutants presented in panel A. Mean values with standard deviation of at least three experiments are shown.
