TABLE 1.
Overview of the simulated distances by different orientations of donor- and acceptor-labeled monomer
All structures are ranked based on the value of sum of squares between the experimental time-resolved fluorescence decay and simulated curves. An interval up to 35% is considered a good fit (green), up to 80% are considered as reasonable (gray), and all values higher than 80% are considered as poor (red) (sum of squares column). Structures are specified according to their basic structural models with specification of the layers in z-direction and the amount of β-strands per monomer. The zipper structures are subdivided in classification with alternating or one-sided orientation of the N termini of each monomer along the z-axis. Color coding in the distances columns indicates the goodness of the fits: best (green, up to 5% deviation), reasonable (gray, up to 25% deviation), or poor (red, above 25% deviation).
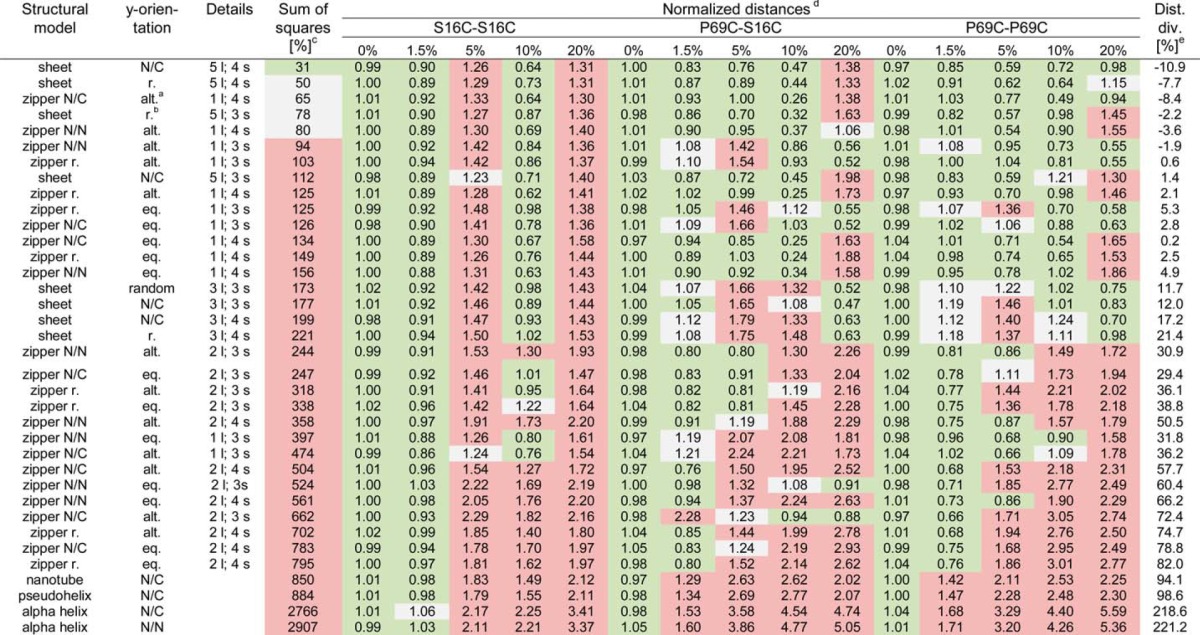
a Abbreviations: alt., alternating; eq. or., equally oriented; r., random; N/N, N to N orientation; N/C, N to C orientation; l., layer; s., strands.
b Note that β-sheet structures with random orientations of the N termini of monomers along the y axis are randomly oriented along the x axis, whereas β-sheet structures with alternating position of the N termini have fixed alignments along the x axis.
c The average sum of squares values represents the excess (in percentage) of the sum of squares as compared with the averaged sum of squares for simulations of 0% acceptor for the given labeling position set.
d Each value is the difference between simulated and experimental photon counts normalized to the averaged difference with 0% acceptor for the same labeling position.
e Dist. div., distance divergence. For each simulated curve, we show the percentages by which the distance between predicted and measured photon amounts vary relative to the average of all fibrils without any acceptor.
