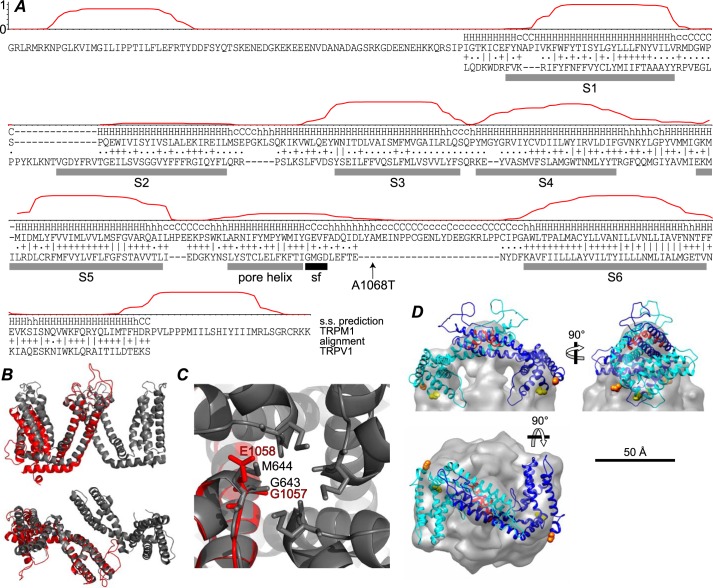
FIGURE 10.
Alignment with TRPV1 and homology model. A, the posterior probability of location in a TM helix, calculated using TMHMM (45), is plotted for residues 784–1176 of TRPM1. The HHpred (46) secondary structure (s.s.) prediction and alignment with TRPV1 is shown below. The ∣, +, and . represent very good, good, and poor matches, respectively. The rat TRPV1 sequence from the structure lacking pore loop residues 604–626 (Protein Data Bank code 3J5P (30)) was used. TRPV1 TM helices S1–S6 and pore helix (gray bars) and selectivity filter (sf) (black bar) are indicated. The location of the TRPM1 A1068T nob mutation (24) is also indicated. B, side view (top) and extracellular view (bottom) of two C2-related (diagonally opposite) TRPV1 TM domains (gray) superposed with the TRPM1 homology model (red) aligned at the S5 helix. C, extracellular view of the TRPV1 tetramer pore region (gray) and TRPM1 homology model (red). TRPV1 selectivity filter residues Gly-643 and Met-644 are indicated as sticks along with candidate TRPM1 selectivity filter residues Gly-1057 and Glu-1058. D, two subunits of the TRPM1 TM domain homology model were fit into the EM map. The N and C termini of the model are shown as orange and yellow spheres, respectively, and the pore helix is red.