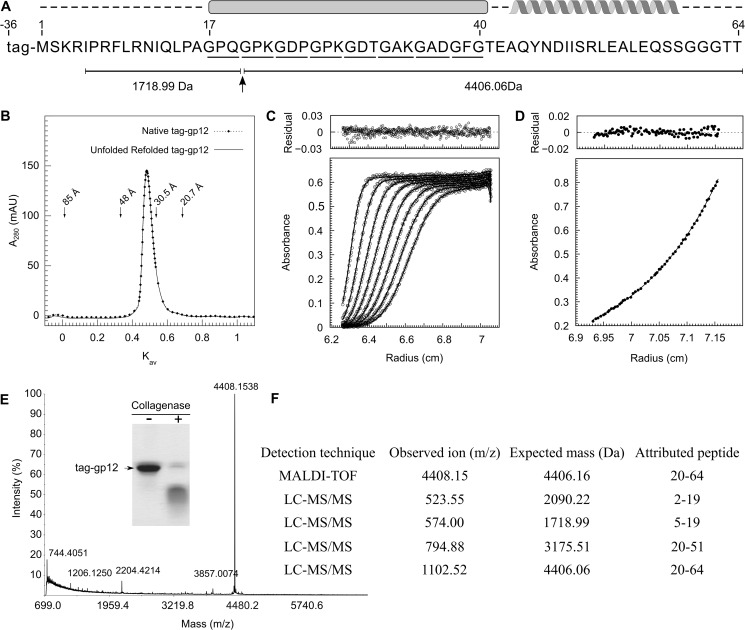
FIGURE 2.
Properties of the SPP1 auxiliary protein gp12. A, Gp12 amino acid sequence showing position of the tag fused to its amino terminus. GXY triplets are underlined. The intermolecular collagen-like triple helix and α-helix predicted by bioinformatics are shown above the sequence. The collagenase cut of tag-gp12 inside the collagen-like sequence motif and peptides obtained from mass spectrometry analysis (E and F) are shown below. B, RH determination of native and unfolded-refolded tag-gp12. Native (dotted line) and tag-gp12 heated for 5 min at 90 °C and transferred directly to ice (continuous line) were analyzed by SEC at 16 °C as described under “Experimental Procedures.” The elution positions of thyroglobulin (RH = 85 Å), γ-globulin (RH = 48 Å), ovalbumin (RH = 30,5 Å), and myoglobin (RH = 20,7 Å) used to calibrate the column are indicated by arrows. Kav (partition coefficient) was calculated as described (35). mAU, milli absorbance units. C, sedimentation velocity of tag-gp12 at 220,000 × g (loading concentration of 1 mg/ml, 16 °C run). Gp12 has a sedimentation coefficient of 1.7 S (s20,w = 2.2 S). D, sedimentation equilibrium of tag-gp12 at 16,300 × g (loading concentration of 0.8 mg/ml, 16 °C run). The data (dots) were fit using a trimer model (continuous line). The best fit was obtained for a single species with an average mass of 31,270 ± 590 Da. The top panels in C and D show the deviation of experimental points from fitted curves. E and F, cleavage of tag-gp12 with collagenase VII analyzed by SDS-PAGE (inset in E) and mass spectrometry. The observed ion mass (4406.15 Da) in MALDI-TOF (E) is attributed to peptide 20–64 of the gp12 sequence, identifying the proteolysis site shown in A. The same peptide was detected by LC-MS/MS spectrometry, which also showed the presence of three other peptides, resulting from collagenase cleavage at the same position (F). The LS-MS/MS analysis had a tag-gp12 sequence coverage of 89%. For clarity, only the peptides in which one end was generated by the collagenase cleavage are listed in F. Those peptides were absent in the analysis of tag-gp12 not treated with collagenase.