Background: An efficient method is required for converting somatic cells to specific cell fates.
Results: A combination of Pitx2 and miR-200a-3p promotes dental epithelial gene expression in cells.
Conclusion: A two-step method efficiently converts mesenchymal cells to dental epithelial cells.
Significance: A new method to generate dental epithelial cells, which are difficult to isolate, is identified for use in regeneration and repair.
Keywords: Beta-catenin (B-catenin), Epithelial-Mesenchymal Transition (EMT), Gene Expression, MicroRNA (miRNA), Paired-like Homeodomain Transcription Factor 2 (PITX2), Reprogramming
Abstract
Pitx2, Wnt/β-catenin signaling, and microRNAs (miRs) play a critical role in the regulation of dental stem cells during embryonic development. In this report, we have identified a Pitx2:β-catenin regulatory pathway involved in epithelial cell differentiation and conversion of mesenchymal cells to amelogenin expressing epithelial cells via miR-200a. Pitx2 and β-catenin are expressed in the labial incisor cervical loop or epithelial stem cell niche, with decreased expression in the differentiating ameloblast cells of the mouse lower incisor. Bioinformatics analyses reveal that miR-200a-3p expression is activated in the pre-ameloblast cells to enhance epithelial cell differentiation. We demonstrate that Pitx2 activates miR-200a-3p expression and miR-200a-3p reciprocally represses Pitx2 and β-catenin expression. Pitx2 and β-catenin interact to synergistically activate gene expression during odontogenesis and miR-200a-3p attenuates their expression and directs differentiation. To understand how this mechanism controls cell differentiation and cell fate, oral epithelial and odontoblast mesenchymal cells were reprogrammed by a two-step induction method using Pitx2 and miR-200a-3p. Conversion to amelogenin expressing dental epithelial cells involved an up-regulation of the stem cell marker Sox2 and proliferation genes and decreased expression of mesenchymal markers. E-cadherin expression was increased as well as ameloblast specific factors. The combination of Pitx2, a regulator of dental stem cells and miR-200a converts mesenchymal cells to a fully differentiated dental epithelial cell type. This pathway and reprogramming can be used to reprogram mesenchymal or oral epithelial cells to dental epithelial (ameloblast) cells, which can be used in tissue repair and regeneration studies.
Introduction
The epithelial stem cells that control growth of the rodent incisor are located in the cervical loop (a stem cell niche) at the posterior end of the incisor (1). The cervical loop consists of the inner enamel epithelium, outer enamel epithelium, and stellate reticulum cells in the core of the cervical loop and a thin layer of stratum intermedium cells (2, 3). The stem cells in the core of the cervical loop will divide and insert into the basal layer of epithelium, the outer enamel epithelium. They will proliferate as transit amplifying cells and then differentiate to ameloblast cells, which secrete enamel. The labial epithelial stem cells differentiate to ameloblasts, whereas odontoblasts originate from mesenchymal stem cells and secrete dentin on both sides of incisor. A signaling network has been proposed to regulate epithelial stem cell proliferation in the cervical loop stem cell niche (4). These signals originate from mesenchymal tissue derived from the neural crest cells to regulate epithelial stem cells in concert with epithelial signals.
The signaling molecules include fibroblast growth factor (FGF)8, expressed in pre-tooth epithelium and bone morphogenic protein (Bmp)4,3 expressed in intervening epithelium and their interaction and competing actions specify tooth formation (5, 6). FGF and Bmp regulate restricted expression of the homeobox transcription factor Pitx2 that is required for tooth development (7). Wnts comprise a large family of secreted ligands that activate several receptor-mediated pathways (8). The well known canonical Wnt/β-catenin pathway activation causes β-catenin accumulation, nuclear translocation, and transcriptional activation by complexes of β-catenin, Lef/Tcf, and Pitx2 transcription factor family members (8–10). Activation of Wnt/β-catenin signaling initiates the de novo formation of hair follicles, feather buds, mammary placodes, taste buds, and teeth (4, 11–17). Wnt/β-catenin signaling is required for multiple stages of tooth development and dental epithelial cell proliferation and differentiation (14). The Lef-1 transcription factor regulates genes involved in cell proliferation and differentiation. Lef-1 deficiency causes arrested tooth development at the bud stage in mice, and the dental epithelial cells fail to survive (18, 19).
miRs are non-coding small RNAs that regulate gene function post-transcriptionally. Animal miRs are imperfectly paired to the 3′-UTR of target mRNA and inhibit protein production either through destabilization of mRNA or inhibition of translation (20). Tooth development, including epithelium stem cell differentiation, is tightly controlled by miRs and a loss of mature miRs results in the development of supernumerary incisors in the Dicer conditional knock-out mouse (21, 22). miRs control stem cell differentiation in the incisor, and miR depletion causes an expansion and increased proliferation of dental stem cells (21).
The miR-200 family regulates the epithelial-mesenchymal transition (EMT) associated with tumor cell migration, invasion, adhesion, and metastasis (23). The miR-200 family targets and represses the expression of genes involved in this process. These genes include Zeb1, Zeb2, and Jagged1 (23–29). The miR-200 family is selectively expressed in differentiating dental epithelial cells and have low levels of expression in the dental stem cell niche (21, 22, 30). The miR-200 family is comprised of five members, miR-429-200a-200b in one cluster and miR-200c-141 in another cluster located on different chromosomes. We recently reported a Pitx2:miR-200c/141:Noggin pathway regulated Bmp signaling and epithelial cell differentiation during odontogenesis (31). Thus, Pitx2 and miR-200 appear to control the fate of dental stem cells.
There are many protocols used for regeneration therapies to develop fully functioning organs including teeth. Current tooth bioengineering relies on the sequential and reciprocal interactions between neural crest-derived mesenchymal cells and stomadial epithelium, in vitro differentiation of dental epithelial progenitor cells through epithelial-mesenchymal interactions and tooth organ germ bioengineering from molar tooth germ-derived epithelial and mesenchymal cells (3, 32–37). However, for replacement of a functional tooth, these tissues are difficult to obtain and maintain in culture. Mesenchymal stem cells derived from bone marrow and dental pulp stem cells are used to make dental cells and tissues, repair dental structures, and regenerate bone (38–42). Stem cells have great promise in tissue bioengineering studies, but they are difficult to obtain. Additionally, more efficient methods are needed for generating dental cells.
The discovery that fibroblast cells can be converted to induced pluripotent cells by induction of a mixture of transcription factors has lead to the development of cell reprogramming for tissue engineering (43). miRs have also evolved as regulators of gene programs that control cell differentiation and cell fate decisions (44). miRs modulate these functions through positive and negative feedback loops to reinforce cellular decisions (45).
Because dental stem cells are difficult to obtain, culture and propagate as well as producing human epithelial-mesenchymal tooth-forming tissues, we propose a new method using a combination of transcription factor and miRs in a sequential addition to both oral epithelial cells and odontoblast mesenchymal cells to produce amelogenin producing dental epithelial cells.
EXPERIMENTAL PROCEDURES
Expression and Reporter Constructs
The expression plasmids containing the cytomegalovirus (CMV) promoter linked to the mmu-miR-200a and mmu-miR-21 precursor were constructed in pSilencer 4.1 (Ambion). Pitx2, and β-catenin S37A expression plasmids were constructed in pcDNA 3.1 MycHisC (Invitrogen) as described previously (46–49). Pitx2 3′-UTR and Pitx2 mutant 3′-UTR generated by mutagenesis (QuikChange site-directed mutagenesis kit, Agilent Technologies) were directionally cloned into the pGL3 CXCR4 1P (Addgene, plasmid 11310). The 7×TopFlash reporter plasmid was constructed into luciferase vector by inserting seven Lef/Tcf binding sites upstream of the minimal thymidine kinase promoter. The FopFlash reporter, which has the Lef/Tcf binding sites mutated, was also constructed in the luciferase vector (50). The Pitx2c 3kb and Lef-1 promoters have been reported previously (9, 51). All constructs were confirmed by DNA sequencing.
Cell Culture, Transient Transfections, Luciferase, and β-Galactosidase Assays
HEK 293 FT, MDPC, and LS-8 cells were cultured in DMEM supplemented with 5 or 10% FBS and penicillin/streptomycin and transfected by electroporation or Lipofectin. The procedures for transient transfections and luciferase and β-galactosidase assays were described previously (46). Transfected cells were incubated for 48 h. LiCl was added to the appropriate cells at a final concentration of 10 mm 23 h before harvest. The pcDNA3.1 empty vector, pLL3.7, or pSilencer 4.1 negative control vectors were added to equalize the total amount of cotransfected expression vectors. SV-40 or CMV β-galactosidase reporter plasmids were co-transfected in all experiments as a control for transfection efficiency. All plasmids were double-banded CsCl-purified.
Generation of MDPC and LS-8 miR-200a Stable Cell Lines
Fragments containing the miR-200a precursor were amplified by PCR and directionally cloned into the EcoRI site of pLL3.7. Lentivirus was generated by cotransfection of the above construct with packaging plasmids into HEK293T cells, as described previously (31). Blastomycin (5 μg/ml) was added to cells for 5 days, and surviving cells were propagated and subcultured. MDPC and LS-8 cells were transduced and subsequently FACS-sorted for green fluorescent protein (GFP), which is co-expressed on a single transcript with the miR.
Chromatin Immunoprecipitation (ChIP) Analysis
The ChIP analysis was performed as described (52) using the ChIP assay kit (Upstate Biotechnology, Inc.) with the following modifications. LS-8 cells were fed for 24 h, harvested, and plated in 60-mm dishes. Cells were cross-linked with 1% formaldehyde for 10 min at 37 °C the next day. Samples were incubated with PITX2ABC rabbit polyclonal antibody (Capra Science) overnight at 4 °C. Immune complexes were washed consecutively for 5 min with each of the following solutions: low salt immune complex wash buffer and high salt immune complex wash buffer four times, and LiCl immune complex wash buffer and TE buffer twice. An aliquot of the immunoprecipitated DNA from non-transfected cells was used for PCR (32 cycles). All reactions were done at an annealing temperature of 61 °C. Two primers for amplifying the Pitx2 binding site in the miR-200a promoter are as follows: sense, 5′-TTCTTGGCTCTGTATGGGAGA-3′; antisense, 5′-CCCCTCTTGCCTTTTTCAG-3′. All of the PCR products were evaluated on a 1% agarose gel in 1× TBE for appropriate size (175 bp) and confirmed by sequencing. As controls, the primers were used without chromatin, and normal rabbit IgG was used replacing the PITX2 antibody to reveal nonspecific immunoprecipitation of the chromatin. Furthermore, the same set of experiments was done with control primers targeting the distal region of miR-200a promoter lacking putative Pitx2 binding sites. The control primers are as follows: sense, 5′-AGGCAACAGACACCTGCACT-3′; antisense, 5′-GAATGACTGTCTCCCCTCCA-3′.
Mouse Tissue Isolation and Real-time PCR Analyses
The PITX2C cDNA was cloned into the K14 promoter construct (53). We placed the hrGFP (humanized Renilla GFP) gene in the cassette to observe expression in live cells and have observed good expression of PITX2C in transgenic mice by PCR. The PITX2C GFP DNA was excised from the plasmid and used for pronuclear injection. Donor female mice (FVB/NCr), stud male (FVB/NCr), vasectomized male (ICR) and recipient female (ICR) were used in the experiments. Multiple founders were analyzed for transgene expression and crossed to BL6 mice and reevaluated for expression (54). All animals were housed in the Program of Animal Resources of the Institute of Biosciences and Technology and were handled in accordance with the principles and procedure of the Guide for the Care and Use of Laboratory Animals. The Texas A&M Health Science Center, Institutional Animal Care and Use Committee approved all experimental procedures. Wild type (C57BL/6) and K14-PITX2C transgenic mouse mandible, maxilla, and palate tissues were harvested at various developmental time points, and total RNA was prepared for analyses of gene and miR expression. These tissues were harvested using modified procedures for isolating epithelial stem cells and tissue (21, 31, 55–58). After removing the skin from mouse heads, the hemi-mandibles and maxilla were isolated by removing muscle, tendons, ligaments, and bones using a scalpel under a dissecting microscope. The incisors and molars were further dissected from bone. The incisors and molars were incubated in dispase II and collagenase I for 30 min at 37 °C to separate epithelium from mesenchyme. Total RNAs were prepared using the RNeasy minikit from Qiagen. The amount and integrity of the RNA samples were assessed by measurement at 260 and 280 nm and gel analyses. LS-8 (59) or MDPC (60) cells transfected with pLL-scramble or pLL-miR-200a precursor stable cell lines were harvested 48 h after seed. Total RNA was reverse transcribed into cDNA by the iScript Select cDNA synthesis kit (Bio-Rad). Realtime PCR was carried out in a total reaction of 25 μl containing 12.5 μl of iQ SYBR Green Supermix, 0.1 μm forward primer, 0.1 μm reverse primer, 0.25 μl of cDNA template in the MyiQ Singlecolor real-time detection system and analyzed by the MyiQ optical system software (version 2.0, Bio-Rad). β-Actin served as a reference gene for normalization of E-cadherin mRNA levels. Lef-1 primer sequences were 5′-GCAGCTATCAACCAGATCC-3 (forward) and 5′-GATGTAGGCAGCTGTCATTC-3 (reverse); E-cadherin primer sequences were 5′-GCTTCAGTTCCGAGGTCTAC-3′ (forward) and 5′-AGATGCCGCTTCACTTGTGAT (reverse); cyclin D2 primer sequences were 5′-GAGCTGCTGGCCAAGATCAC (forward) and 5′-GACTTGGATCCGGCGTTATG (reverse) (50). Dog E-cadherin primer sequence were 5′-AGTGACTCGCAATGATGTGG-3′ (forward) and 5′-GAACCGCTTCCTTCATAGTC-3′ (reverse) and dog GAPDH primers sequence 5′-CATCACTGCCACCCAGAAG-3′ (forward) and 5′-CAGTGAGCTTCCCGTTCAG-3′ (reverse) (27). The thermal cycling profile consisted of 95 °C for 4 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 72 °C for 18 s. Samples were run in triplicate. No-template control was run in each experiment. Melting curve analyses were performed to confirm amplification specificity of the PCR products, and all PCR products were sequenced to confirm their identity.
Western Blot Assays
Expression of endogenous Pitx2 and β-catenin proteins were demonstrated using the PITX2ABC antibody (Cappa Science) and β-catenin antibodies (Millipore). Approximately 10–40 μg of transfected cell lysates or tissues were analyzed in Western blots. Following SDS gel electrophoresis, the proteins were transferred to PVDF filters (Millipore), immunoblotted, and detected using specific antibodies and ECL reagents from GE Healthcare.
Immunofluorescent Staining
The pLL-Scramble, pLL-Pitx2, pLL-miR-200a, and pLL-Pitx2-miR-200a cells were plated onto fibronectin-coated chamber slides (BD Biosciences) and stained at day 12 or day 3, respectively. For E-cadherin and amelogenin staining, cells were washed with PBS, fixed in ice-cold acetone for 10 min at 4 °C, and air-dried. Fixed cells were washed with PBS for 2–5 min and incubated in 10% normal goat serum-PBS 30 min at room temperature, and then cells were incubated with rat anti-E-cadherin or anti-amelogenin antibodies at 4 °C overnight. After overnight incubation, cells were rinsed by washing in PBS for 3–5 min, and cells were incubated with Alexa Fluor 488 goat anti-rat IgG (H+L) (Molecular Probes, labeling & detection by Invitrogen) 30 min at 37 °C. Cells were washed with PBS for 3–5 min and mounted with VECTASHIELD Mounting Medium with DAPI (Vector Laboratories, Inc., Burlingame, CA). Images were acquired using a Nikon ECLIPSE 80i microscope and NIS-Elements AR software (version 3.2).
RNA Sequencing
Total RNA was isolated from LS-8 and MDPC-23 cell lines after transduction with virus and fluorescence-activated cell sorted (FACS), using the miRNeasy mini kit (Qiagen) following the manufacturer's instructions. RNA quality and concentration was determined using the Agilent 321 Bioanalyzer (Agilent Technologies). Using 1 μg of RNA and the TruSeq Stranded Total RNA Library Prep kit (Illumina), reverse transcription with bar-coded primers, complementary DNA amplification, and 100 × 100 paired ended sequencing with Illumina HiSeq 2000 were performed. Each sample had one biological duplicate that was barcoded separately and sequenced in an independent lane. Quality control of the obtained reads and mapping to the mouse reference genome (GRCm38/mm10) were performed using the combination of the Galaxy web-based analysis suite (61–63) and in-house Perl scripts. Mapped reads were analyzed using Cufflink tool set (64) to identify significant changes in gene expression. The low expression transcripts (<10 reads in all samples) were filtered out, and p values were adjusted using a threshold for false discovery rate ≤ 0.001. Differentially expressed transcripts were identified using threshold of fold change ≥ 2 and false discovery rate ≤ 0.001. The differentially expressed genes were further used for hierarchical clustering performed using Cluster (version 3.0), and Java Treeview was used for visualization (65, 66). Gene ontology category enrichment was assessed using GOrilla. Q-RT-PCR analysis for the genes indicated was compared with the RNA sequencing results. Amplification of VPS29 and actin was used to normalize the values.
RESULTS
Bioinformatics analyses of isolated murine dental epithelium and mesenchyme at different developmental stages identified gene and miR signatures. These data were then integrated into gene ontology category enrichment maps and used to identify processes that are common and different for dental epithelial cells compared with other cells. These data identified several new molecular mechanisms that control dental epithelial cell proliferation and differentiation. Pitx2 is the first transcriptional marker for tooth development, and the miR-200 family is required for dental epithelial cell differentiation (6, 21, 31, 67, 68). We first performed a functional analyses of a new Pitx2-β-catenin-miR-200a regulatory pathway identified in the bioinformatics analyses and then used this new mechanism to convert both oral epithelial and dental mesenchymal cells to a dental epithelial cell fate. We reported a method to convert oral epithelium and dental mesenchymal cells to dental epithelial cells, which express many genes that are hallmarks of these cells, including the expression of the ameloblast (differentiated dental epithelium)-specific marker, amelogenin.
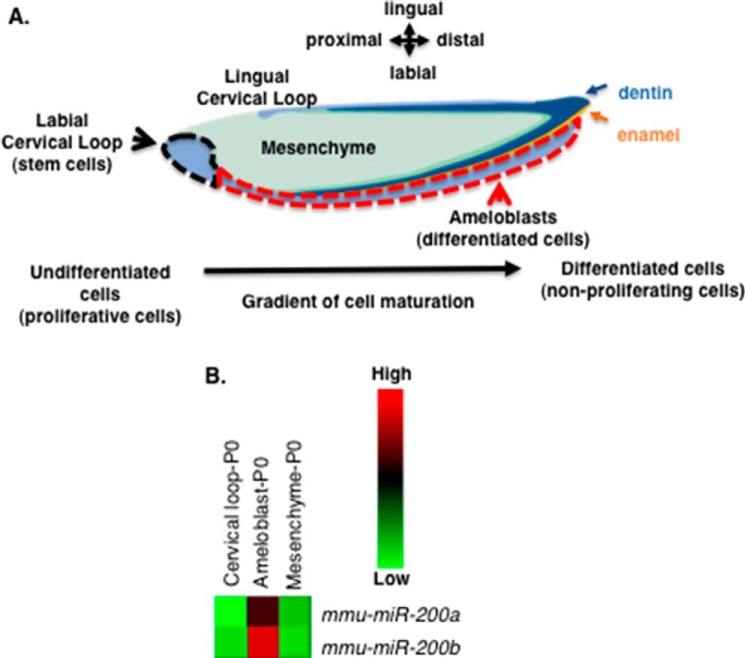
Dental epithelial and mesenchymal tissue from P0 mouse incisors were isolated and screened for gene and miR expression to understand gene regulatory networks (GRNs) and how dental stem cells yield fully differentiated epithelial cells or ameloblasts. The incisor labile cervical loop (LaCL) contains dental stem cells that give rise to the differentiated ameloblasts (Fig. 1A). The epithelial compartment was divided into the LaCl or undifferentiated cells (black dashed line) and the differentiating cells (red dashed line), whereas the dental mesenchyme was separated from the epithelium, and total RNA was isolated for gene and miR expression analyses. A miR expression profile was identified showing that miR-200a and miR-200b were associated with differentiating epithelial cells (pre-ameloblast cells) (Fig. 1B). These miRs were not expressed in the dental mesenchyme, suggesting that miR-200a and miR-200b may promote epithelial cell differentiation. However, their molecular mechanism in dental epithelial cell differentiation was not known.
FIGURE 1.
miR-200a-3p expression is associated with differentiating dental epithelial cells. A, schematic of the mouse lower incisor cell and tissue structures. The black dotted line denotes the labial cervical loop (LaCL, stem cell niche), the red dotted line denotes the pre-secretory, secretory, and mature differentiated epithelial tissues (pre-ameloblasts and ameloblasts). Green shaded region, mesenchyme; dark blue, dentin; orange, enamel. B, heat map of selected miR-200a-3p and miR-200b-3p expression in the isolated dental epithelial tissue compartment (LaCL versus ameloblasts) and dental mesenchyme (ameloblast versus mesenchyme). These tissues were isolated from P0 mice lower incisors, total RNA was harvested, and miRs were analyzed by microRNA arrays. Five separate biological samples were analyzed. mmu, Mus musculus.
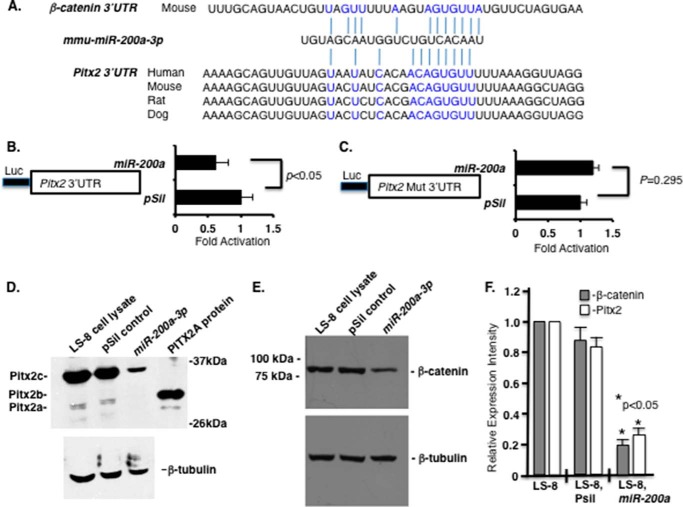
Analyses of miR-200a-3p binding elements identified Pitx2 and β-catenin as potential targets through sequences in their 3′-UTRs (Fig. 2A). To demonstrate a functional miR-200a-3p regulation of Pitx2, the Pitx2 3′-UTR was cloned into the luciferase vector and co-transfected in LS-8 oral epithelial cells with miR-200–3p or empty vector. miR-200a-3p repressed luciferase activity from the vector containing the Pitx2 3′-UTR, but not when the miR-200a-3p target site was mutated in the Pitx2 3′-UTR construct (Fig. 2, B and C). Transfected miR-200a-3p decreased endogenous Pitx2 protein expression (Pitx2 isoforms) in LS-8 cells (Fig. 2D) and also decreased β-catenin protein expression in these cells (Fig. 2E) (69). Quantitation of multiple Western blots demonstrates significantly decreased Pitx2 and β-catenin protein expression in miR-200a transfected LS-8 oral epithelial cells (Fig. 2F). These data demonstrate that both Pitx2 and β-catenin are targeted by miR-200a-3p.
FIGURE 2.
miR-200a-3p directly targets the Pitx2 and β-catenin 3′-UTR and represses Pitx2 and β-catenin expression. A, miR-200a-3p is evolutionarily conserved among several vertebrate species, and the miR-200a target sites in the Pitx2 and β-catenin 3′-UTR are shown. B, Pitx2 3′-UTR luciferase construct transfected with either miR-200a or empty vector in LS-8 cells. Luciferase (Luc) activity was measured using the Dual-Luciferase system to control for transfection efficiency and normalization (n = 3; p < 0.05). C, as a control, the miR-200a target site was mutated in the Pitx2 3′-UTR and showed no inhibition when co-transfected with miR-200a or empty vector (n = 3; p = 0.295). D, miR-200a-3p represses endogenous Pitx2a and Pitx2c isoform expression. Western blot of endogenous Pitx2 in miR-200a precursor transfected LS-8 cells 48 h post-transfection. β-Tubulin is shown as a loading control. The Pitx2b isoform was not detected in LS-8 cells. E, miR-200a represses endogenous β-catenin expression. Western blot of β-catenin protein in control or miR-200a precursor transfected LS-8 cells 48 h post-transfection. β-Tubulin is shown as a loading control. LS-8 cell lysate (empty vector, mock) and pSil-neg vector served as controls. F, quantitation of β-catenin and Pitx2 endogenous expression from three Western blots using different LS-8 cell lysates expressing miR-200a or controls (empty vector or no vector; n = 3; *, p < 0.05). mmu, Mus musculus.
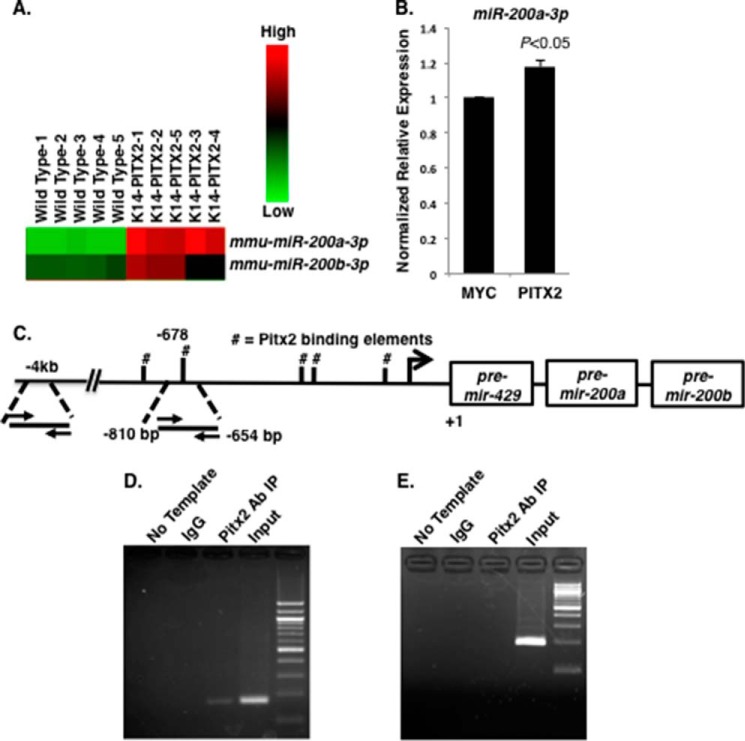
Mouse incisor P1 epithelial tissue from wild type (WT) and K14-PITX2C overexpression mice was analyzed for miR expression by miR arrays. PITX2C overexpression increased miR-200a and miR-200b expression in the dental epithelium (Fig. 3A). Sequence analyses of the 5′-flanking region of the miR-429-200a-200b cluster identified a Pitx2-binding element (5′-TAATCC-3′). We next asked whether Pitx2 regulated miR-200a-3p expression. Transfection of Pitx2C in LS-8 cells activated endogenous miR-200a-3p expression (Fig. 3B). Endogenous Pitx2 binding to the miR-200a promoter was characterized by ChIP in LS-8 cells. Pitx2 bound to a Pitx2 element in the 5′-flanking region of the miR-200a chromatin (Fig. 3C) but not to an adjacent sequence. This sequence does not contain a Pitx2 binding site (Fig. 3, D and E, respectively). Thus, Pitx2 activates miR-200a-3p expression and miR-200a-3p acts to repress Pitx2 expression. This tight regulation of Pitx2 and miR-200a expression may be one mechanism to control dental epithelial differentiation.
FIGURE 3.
Endogenous Pitx2 binds to and activates the miR-200a promoter. A, heat map of miR-200a-3p and miR-200b-3p expression in wild type and K-14-PITX2 overexpression mouse lower incisor epithelial tissue. B, real time PCR of endogenous miR-200a-3p expression in LS-8 cells transfected with empty vector (Myc) or PITX2 (n = 3). C, schematic representation and location of the Pitx2-binding site in the mmu-miR-429-200a-200b promoter. #, the Pitx2 binding elements (TAATCC). D, ChIP of endogenous Pitx2 binding to the Pitx2 element ∼678 bp upstream of pre-miR-429-200a-200b transcript in LS-8 cells. Rabbit antisera were used as a control IP and Pitx2ABC antisera from CAPPA SCIENCE was used to IP Pitx2 binding to the chromatin. The input chromatin is shown as a positive control for the ChIP. E, control ChIP using the Pitx2 antisera and primers to a 4-kb upstream region of the pre-miR-429-200a-200b transcript. This chromatin does not contain a Pitx2-binding site and was not immunoprecipitated using Pitx2 antisera; the primers did amplify the input chromatin.
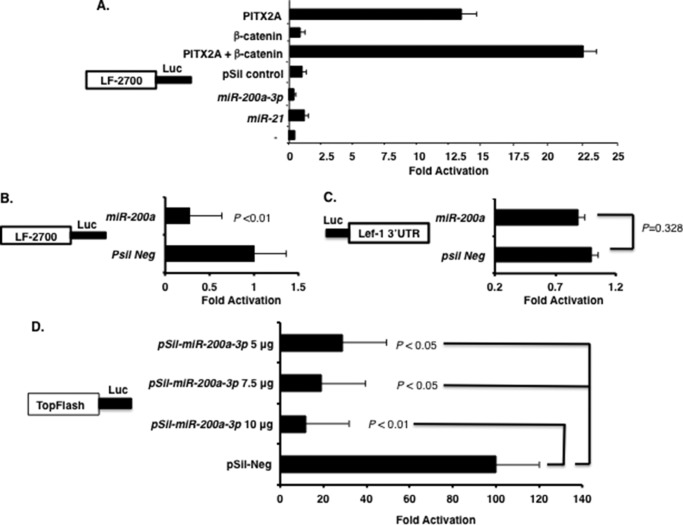
Wnt/β-catenin expression is required for tooth development at many stages and regulates incisor epithelial cell proliferation and differentiation (14). Pitx2 activates Lef-1 expression and in concert with β-catenin synergistically activates the LEF-1 promoter (Fig. 4A) (9). When LS-8 cells (which endogenously express Pitx2 and β-catenin) were cotransfected with the LEF-1 promoter and miR-200a, luciferase activity from the LEF-1 promoter decreased compared with miR empty vector and miR-21 (used as controls) (Fig. 4A). A direct comparison of LEF-1 promoter activity with miR-200a expression revealed a 4-fold decrease in luciferase activity compared with empty vector (Fig. 4B). As a control, we show that miR-200a does not regulate the Lef-1 3′-UTR luciferase construct (Fig. 4C). Thus, miR-200a-3p indirectly regulates Lef-1 expression by inhibiting endogenous Pitx2 and β-catenin, which both activate endogenous Lef-1 expression. Titration of miR-200a-3p in LS-8 cells transfected with the TopFlash reporter decreased its luciferase activity compared with empty vector alone (Fig. 4D). The FopFlash reporter (LEF-1 binding sites mutated) was not affected by miR-200a-3p expression (data not shown). Thus, Pitx2 and β-catenin interact (9, 10) to regulate Lef-1 expression and miR-200a-3p represses Pitx2 and β-catenin expression and indirectly represses Lef-1 expression and Wnt/β-catenin signaling mechanisms.
FIGURE 4.
miR-200a-3p indirectly regulates the LEF-1 promoter and TopFlash reporter. A, miR-200a targets endogenous Pitx2 and β-catenin, which activate the LEF-1 promoter in LS-8 cells. The LEF-1 2.7-kb mouse promoter and Pitx2, β-catenin, pre-miR-200a, pSil-empty vector, and pre-miR-21 were transfected in LS-8 cells. To control for transfection efficiency, all transfections included the SV-40 β-galactosidase reporter (0.5 μg). Cells were incubated for 48 h and then assayed for luciferase (Luc) and β-galactosidase activities as described previously (10). The activities are shown as mean fold activation compared with the luciferase plasmid with empty vector and normalized to β-galactosidase activity ± S.E. from three independent experiments. B, miR-200a targets endogenous Pitx2 and represses Lef-1 activity in LS-8 cells. The LEF-1 promoter was transfected with miR-200a or pSil vector only in LS-8 cells, which endogenously express Pitx2 and β-catenin. Luciferase activity was assayed, and miR-200a transfection was compared with vector only as in A. C, as a control, the Lef-1 3′-UTR luciferase construct was transfected with miR-200a or empty vector to demonstrate that miR-200a does not directly regulate Lef-1 expression. D, the TopFlash reporter (contains seven Lef-1 binding elements (50)) was co-transfected with increasing amounts of miR-200a-3p plasmid in LS-8 cells. Luciferase activity was measure as described in A.
Pitx2 is highly expressed in the LaCL and regulates progenitor cell proliferation and differentiation during incisor development along with Lef-1 (7, 14, 18, 31). Because miR-200a caused an MET response when overexpressed in cells (27), we asked whether miR-200a-3p could cause the LS-8 oral epithelial-like cells to convert to dental epithelial cells that express E-cadherin. Furthermore, E-cadherin, which promotes epithelial cell differentiation also decreases β-catenin transcriptional activity (70). However, this E-cadherin repression of β-catenin can be counteracted by Pitx2 activation of Lef-1 expression. Thus, miR-200a induces E-cadherin expression and MET, whereas Pitx2 acts to increase Lef-1 expression and cell proliferation.
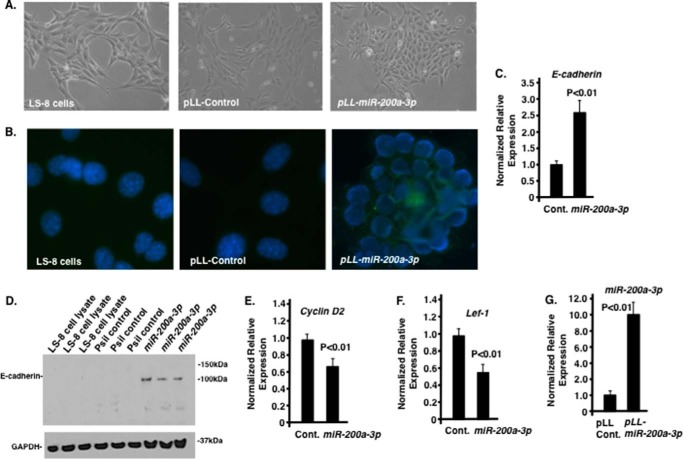
LS-8 cells transduced with lentiviral scrambled or miR-200a-3p constructs were assayed for gene expression and cell morphology changes after 2 weeks in culture. The LS-8 cells grow similar to fibroblast cells in culture (59) and when transduced with scrambled control vector (Fig. 5A). After miR-200a transduction (pLL-miR-200a) these cells form clusters of cells that more closely resemble epithelial cells (Fig. 5A). Immunocytochemistry using an E-cadherin antibody showed increased E-cadherin expression in miR-200a transduced cells (Fig. 5B). Gene expression analyses of the miR-200a-3p transduced LS-8 cells showed increased E-cadherin (both transcripts and protein, Fig. 5, C and D) and miR-200a expression and decreased cyclin D2 and Lef-1 expression compared with scrambled control cells (Fig. 5, E–G).
FIGURE 5.
Effect of miR-200a-3p expression in LS-8 cells. A, morphology change of LS-8 cells transduced with pLL-control and pLL-miR-200a lentivirus constructs. B, E-cadherin staining (green fluorescence) of LS-8, pLL control, and pLL-miR-200a transduced cells. DAPI staining used to detect nuclei. C and D, quantitation by real time PCR and Western blot of E-cadherin in LS-8 cells transfected with pLL control (Cont.) vector and PLL-miR-200a, respectively. E and F, quantitation by real time PCR of cyclin D2 and Lef-1 in LS-8 cells transfected with pLL control vector and PLL-miR-200a, respectively. G, as a control, miR-200a expression was increased in pLL-miR-200a-3p-transduced LS-8 cells.
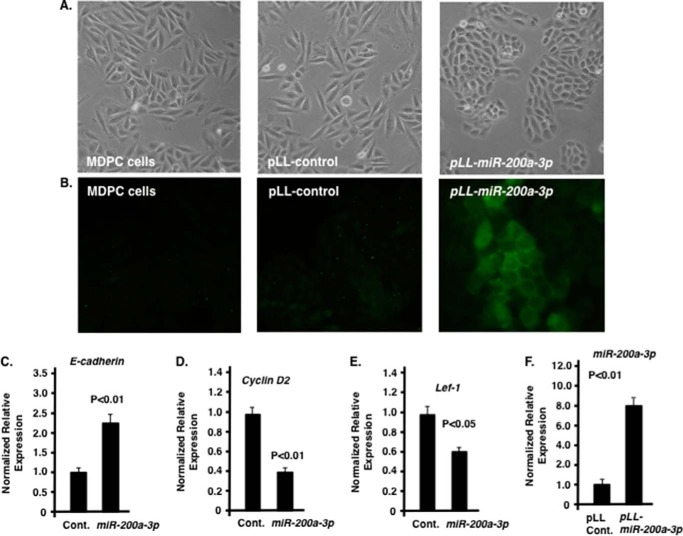
The MDPC odontoblast mesenchymal cells (60) were also transduced with scrambled and miR-200a-3p lentiviral constructs and the cells expressing miR-200a-3p showed an epithelial cell morphology (Fig. 6A). The MDPC cells transduced with miR-200a-3p expressed high levels of E-cadherin in culture (Fig. 6B). The transduced MDPC cells have increased E-cadherin and miR-200a-3p expression and decreased cyclin D2 and Lef-1 expression similar to the miR-200a transduced LS-8 cells (Fig. 6, C–F). Interestingly, miR-200a-3p overexpression alone was not sufficient to induce a GRN associated with dental epithelial cells (data not shown). Bioinformatics analyses of both LS-8 and MDPC cells transformed with miR-200a did not activate the dental epithelial cell program identified in mouse dental epithelial tissue.
FIGURE 6.
Effect of miR-200a-3p expression in MDPC cells. A, morphology change of MDPC cells transduced with pLL-control and pLL-miR-200a-3p. B, E-cadherin staining (green fluorescence) of MDPC, pLL control, and pLL-miR-200a transduced cells. C–E, quantitation by real time PCR of E-cadherin, cyclin D2, and Lef-1 in MDPC cells transduced with pLL control (Cont.) vector or PLL-miR-200a. F, as a control, miR-200a expression was increased in pLL-miR-200a-3p-transduced MDPC cells.
Several combinations of transcription factors and miRs were analyzed for their effect to reprogram these cells. However, Pitx2 and miR-200a were the best candidates. We first transduced both cell types with Pitx2, and cells were treated with blastomycin to select stable transduced cells. After 4 weeks, the cells were transduced with miR-200a that express GFP, and stable cells were initially selected using blastomycin treatment, and after culture for 1 week, the cells were FACS sorted using GFP as the marker. Two weeks later, the cells were analyzed for morphology and gene expression.
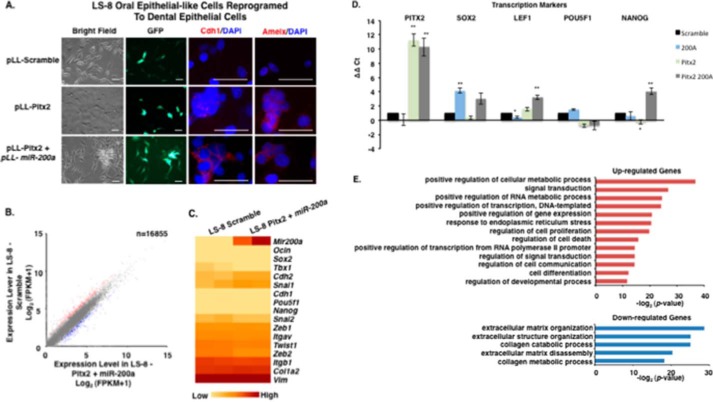
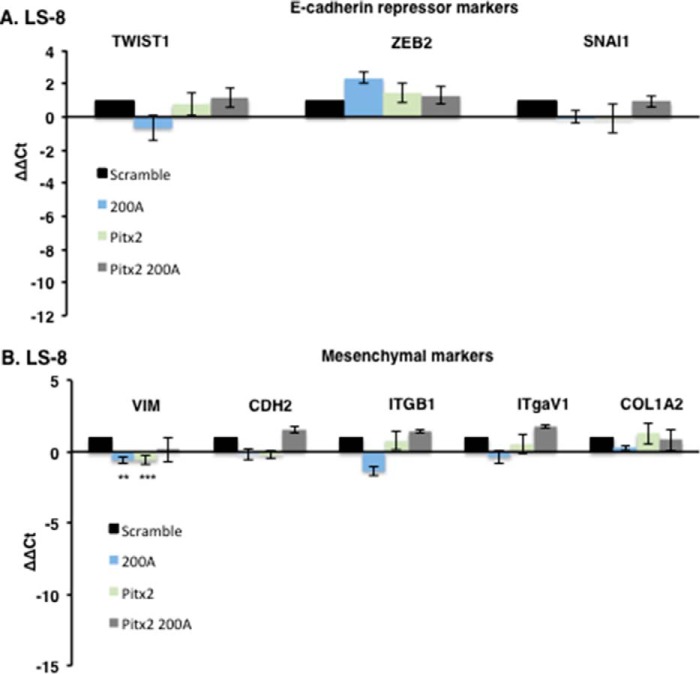
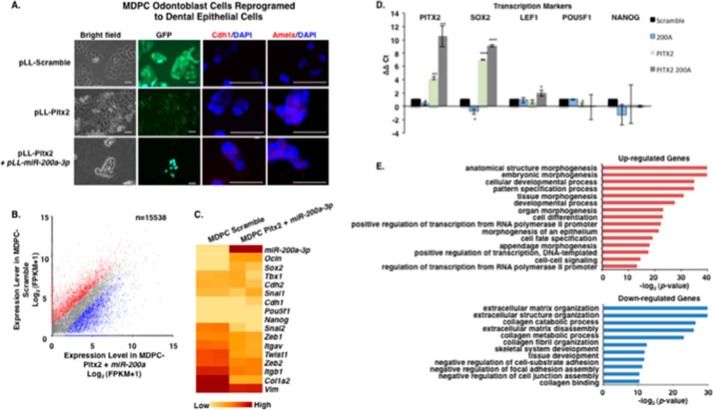
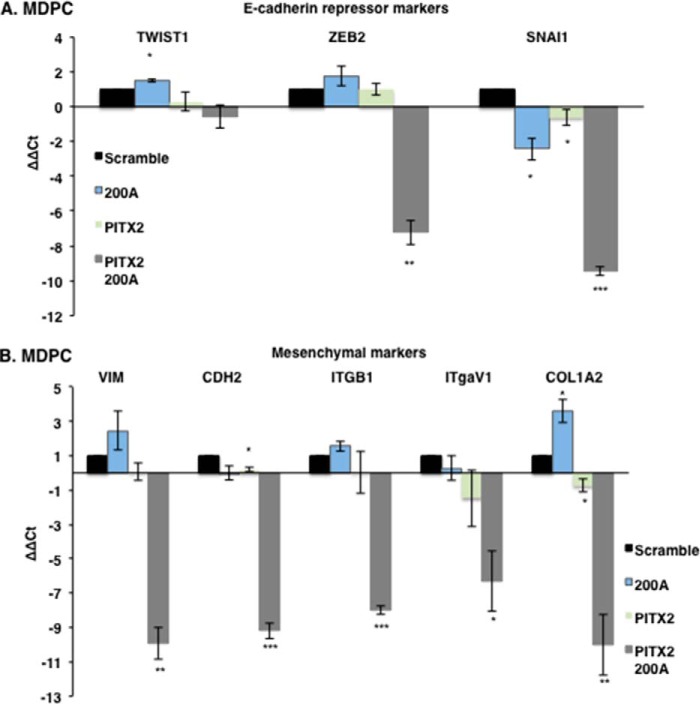
E-cadherin and amelogenin are well known dental epithelial differentiation markers and characterize fully differentiated ameloblast cells (71–76). LS-8 cells after Pitx2 transduction express amelogenin (Amelx) but not E-cadherin (CDH1) (Fig. 7A); it has been shown that increased Lef-1 expression reduces E-cadherin expression (70). However, after transduction with Pitx2 and miR-200a, both E-cadherin and amelogenin were expressed (Fig. 7A, bottom panel). Gene expression involved in MET in the Pitx2 and miR-200a transduced LS-8 cells was analyzed by RNA-seq and confirmed by quantitative PCR (Fig. 7, B–E). Real time PCR confirmed the up-regulation of Pitx2, Sox2, Lef-1, and Nanog in converted LS-8 cells (Fig. 7D) and genes that regulate transcription and signal transduction from the RNA-seq experiment (Fig. 7E). Conversely, in LS-8 reprogrammed cells E-cadherin repression markers and mesenchymal markers were not changed significantly compared with scrambled control cells (Fig. 8, A and B). RNA-seq identified multiple genes associated with collagen and extracellular matrix processes that were down-regulated (Fig. 7E). These data are consistent with conversion of LS-8 oral epithelial cells to dental epithelial cells. A selected list of dental epithelial (pre-ameloblast and ameloblast) transcription factors and other genes are listed in Table 1. These genes are highly expressed and appear to be signature genes for differentiated dental epithelium. We compare the converted LS-8 oral epithelial cells and MDPC cells to this signature. Most of these factors are expressed in the oral epithelial cells, and we find modest increases in their expression after Pitx2 and miR-200a expression. However, the converted MDPC dental mesenchymal cells demonstrate large increases in the expression of these dental epithelial genes.
FIGURE 7.
Reprogrammed LS-8 oral epithelial cells express amelogenin and dental epithelial factors. A, LS-8 oral epithelial cells are transduced with pLL-Pitx2 and a combination of pLL-Pitx2 and pLL-miR-200a or lentiviral vector expressing a pLL-scrambled RNA control. Cells were FACS sorted, and GFP and immunofluorescence microscopy analysis of changes in the expression levels of Cdh1 and Amelx were observed after 8 weeks. Pitx2-transduced cells express low levels of E-cadherin (Cdh1) and amelogenin (Amelx). Pitx2 and miR-200a-transduced cells express both Cdh1 and Amelx and form tight junctions between cells. GFP expression shows the cells were transduced with the lentiviral vector. B, RNA-seq analysis of gene expression in response to Pitx2-miR-200a overexpression in LS-8 cells. Significantly up- and down-regulated genes that have at least a 2-fold of expression level change were labeled red and blue, respectively. All expression levels were estimated by FPKM. C, heat map showing the expression dynamics of selected EMT genes upon Pitx2-miR-200a overexpression. Epithelial specific and mesenchymal specific genes were hierarchically clustered, respectively. D, real time PCR of selected transcription factors associated with dental epithelial proliferation and differentiation. Endogenous Sox2, Lef-1, and Nanog were increased in the Pitx2-miR-200a-transduced LS-8 cells. Pitx2 was overexpressed as expected. (Pitx2 cDNA is not regulated by miR-200a.) Pou5f1 (Oct4) was not changed significantly (n > 3; *, p < 0.05, **, p < 0.01). E, gene ontology (GO) analysis significantly up- and down-regulated genes. Top enriched GO terms (−log2 p value > 10) are highlighted for both up-regulated (red) and down-regulated (blue) genes that are related to EMT and morphogenic functions. FPKM, fragments per kilobase of transcript per million mapped reads.
FIGURE 8.
FACS sorted transduced LS-8 cells were analyzed for specific gene expression. A, the E-cadherin repressor genes Twist1, Zeb2, and Snail1 were all increased in the Pitx2-miR-200a-transduced cells (n = 3). B, mesenchymal markers were not increased above scrambled control cells, and the expression levels were low (>Ct 30; n = 3).
TABLE 1.
Selected murine P0 dental epithelial markers were identified by RNA-seq and DNA microarrays and compared with dental mesenchyme gene expression
RNA-seq experiments identified gene expression changes of the epithelial cell markers in converted oral epithelial (epi.) cells and dental mesenchyme (mes.) cells. Shown are selected murine dental epithelial genes in P0 mouse dental epithelium compared with mesenchymal cells; converted oral epithelial (LS-8) cells compared with non-transformed oral epithelial (LS-8) cells; and converted dental mesenchymal (MDPC) cells compared with non-transformed dental mesenchymal (MDPC) cells. exp., expression.
| Gene | Expression levels | P0 dental mes. vs. P0 dental epi. | Converted cells vs. oral epi. cells | Converted cells vs. dental mes. cells |
|---|---|---|---|---|
| Tbx5 | High exp. | (−) 15-fold | (+) 2-fold or less | (+) 4.1-fold |
| Satb1 | Med. exp. | ( − ) 2 − fold | (+) 2-fold or less | (+) 10-fold |
| Pitx1 | High exp. | (−) 7-fold | (+) 2-fold or less | (+) 18-fold |
| Lhx6 | Med. exp. | (−) 8.5-fold | (+) 2-fold or less | (+) 2.1-fold |
| Isl1 | High exp. | (−) 6.3-fold | (+) 2-fold or less | (+) 10.4-fold |
| Dach1 | Med. exp. | (−) 2.7-fold | (+) 2-fold or less | (+) 7.9-fold |
| Atf2 | Med. exp. | (−) 4.1-fold | (+) 2-fold or less | (+) 2.8-fold |
| Lef1 | Med. exp. | (−) 2.4-fold | (+) 2-fold or less | (+) 15.4-fold |
| Hmx1 | Low exp. | (−) 3.2-fold | (+) 2-fold or less | (+) 10.8-fold |
| Tbx1 | High exp. | (−) 5.5-fold | (+) 2-fold or less | (+) 2-fold or less |
| Foxj3 | High exp. | (−) 10-fold | (+) 2-fold or less | (+) 6.2-fold |
| Irf6 | High exp. | (−) 7-fold | (+) 2-fold or less | (+) 2-fold or less |
| Cacna1g | High exp. | (−) 9-fold | (+) 1.4-fold | (+) 2-fold or less |
| Gadd45a | High exp. | (−) 2.6-fold | (+) 2.3-fold | (+) 2-fold or less |
| Mmp20 | Low exp. | (−) 1.6-fold | (+) 2-fold or less | (+) 2.1-fold |
| Enam | High exp. | (−) 11-fold | (+) 2-fold or less | (+) 2-fold or less |
MDPC mesenchymal cells after miR-200a transduction express E-cadherin (CDH1) but not amelogenin; however, after Pitx2 transduction, these cells express amelogenin but not E-cadherin (Fig. 9A). Transduction with Pitx2 and miR-200a stimulated both E-cadherin and amelogenin expression (Fig. 9A). Gene expression involved in MET in the Pitx2 and miR-200a transduced MDPC cells was analyzed by RNA-seq and confirmed by quantitative PCR (Fig. 9, B–E). Real time PCR confirmed the up-regulation of Pitx2, Sox2, and Lef-1 in converted MDPC cells, and the RNA-seq experiments identified genes that regulate transcription and signal transduction (Fig. 9, D and E). As expected, in MDPC converted cells, E-cadherin expression was up-regulated, and mesenchymal markers were all down-regulated (Fig. 10, A and B). The significant gene expression changes demonstrate that these reprogrammed MDPC cells have undergone an epithelial conversion to a dental epithelial cell fate. RNA-seq identified multiple genes that were down-regulated and associated with collagen and extracellular matrix processes indicative of mesenchymal cells (Fig. 9E).
FIGURE 9.
Reprogrammed MDPC odontoblast cells express amelogenin and dental epithelial factors. A, MDPC mesenchymal cells are transduced with Pitx2 and a combination of Pitx2 and miR-200a or lentiviral vector expressing a scrambled RNA control. Cells were FACS sorted, and GFP and immunofluorescence microscopy analysis of changes in the expression levels of Cdh1 and Amelx were observed after 8 weeks. Pitx2-transduced cells express low levels of E-cadherin (Cdh1) and amelogenin (Amelx). Pitx2 and miR-200a-transduced cells express both Cdh1 and Amelx and form tight junctions between cells. GFP expression shows the cells were transduced with the lentiviral vector. B, RNA-seq analysis of gene expression in response to Pitx2-miR-200a overexpression in MDPC cells. Significantly up- and down-regulated genes that have at least 2-fold expression level change were shaded red and blue, respectively. All expression levels were estimated by FPKM. C, heat map showing the expression dynamics of selected EMT genes upon Pitx2-miR-200a overexpression. Epithelial specific and mesenchymal specific genes were hierarchically clustered, respectively. D, real time PCR of selected transcription factors associated with dental epithelial proliferation and differentiation. Endogenous Sox2 and Lef-1 were increased in the Pitx2-miR-200a transduced LS-8 cells. Pitx2 was overexpressed as expected (Pitx2 cDNA not regulated by miR-200a). Pou5f1 (Oct4) and Nanog were not significantly changed (n > 3; *, p < 0.05; **, p < 0.01). E, gene ontology (GO) analysis significantly up- and down-regulated genes. Top enriched GO terms (–log2 p value > 10) are highlighted for both up-regulated (red) and down-regulated (blue) genes that are related to EMT and morphogenic functions.
FIGURE 10.
FACS sorted transduced MDPC cells were analyzed for specific gene expression. A, the E-cadherin repressor genes Zeb2 and Snail1 were significantly decreased in Pitx2-miR-200a transduced cells compared with scrambled control (n = 3). B, all measured mesenchymal markers were significantly down-regulated in Pitx2-miR-200a transduced cells compared with scrambled control (n = 3).
DISCUSSION
Recently, there has been an explosion of studies using dental stem cells and isolated dental epithelial-mesenchymal interactions to generate epithelial cells and tissue for tooth bioengineering and regeneration (1, 59, 74, 77–89). Many of the genes required for epithelial cell proliferation and differentiation during tooth organogenesis and regeneration have been identified and are being used in research to make teeth (33–35, 90). There are an abundance of studies using dental stem cells, mesenchymal stem cells, bone marrow stem cells, and epithelial-mesenchymal interactions to differentiate epithelial cells capable of regenerating dental epithelium and dental structures (>100 manuscripts published). A common theme in these studies relies on the isolation of dental progenitor or stem cells to generate competent differentiated dental epithelial cells. These procedures are intrusive and provide limited amounts of material. A procedure to convert easily obtained oral epithelial cells or dental mesenchymal cells from patients would greatly facilitate tooth and tissue regeneration.
miRs have been identified as key regulators of progenitor cell differentiation and modulators of cell fate decisions (21, 31, 44, 45, 91). miRs regulate the fate of stem cells in many different tissues and organs through the specification or differentiation of cell types. miRs can target cell cycle regulators, promote differentiation by inactivating transcriptional repressors, and integrate with transcriptional and signaling networks in bone formation, muscle differentiation, neurogenesis, and tooth and craniofacial morphogenesis (21, 31, 44, 45, 92). The use of miRs in cell reprogramming is a new field of research that has great promise for tooth regeneration.
Wnt/β-catenin signaling, Tcf/Lef-1, and Sox factors control stem cell renewal and differentiation in many tissues and organs, including teeth (14, 17, 93–95). Lef-1 is required for early tooth formation and cell proliferation (18, 19). Pitx2 also controls dental progenitor cell proliferation and differentiation (31, 96). Pitx2 physically interacts with β-catenin to activate Lef-1 expression and miR-200c expression to inhibit Noggin and activate BMP to allow for dental epithelial cell differentiation (9, 10, 31). This unique signaling network and GRN suggested that these factors might regulate dental epithelial cell fate. Our bioinformatics analyses demonstrate that Wnt signaling-associated factors involved in odontogenesis are increased in Pitx2-miR-200 converted cells, including Fzd9, Fzd8, Wnt11, and Fzd6. Furthermore, we constructed a list of highly expressed factors expressed in P0 mouse dental epithelium, which we use as a signature to designate dental epithelium (Table 1). This signature only denotes the highly expressed genes and excludes other genes (not highly expressed) that are also associated with dental epithelium. The converted dental mesenchyme to dental epithelium cells all highly express these factors. Interestingly, the oral epithelial cells express these factors and thus are only moderately increased in the converted dental epithelial cells.
In this report, we have identified a new GRN where Pitx2 activates miR-200a expression, and this miR then feeds back to modulate Pitx2 expression as dental progenitor cells differentiate. miR-200a represses β-catenin expression and this GRN then indirectly modulates Lef-1 expression. miR-200a is a regulator of dental epithelial progenitor cell differentiation. However, miR-200a can also specify epithelial cell fates by repressing Zeb expression to allow for E-cadherin expression. This unique molecular mechanism guides tissue morphogenesis. Here, we hijacked this process by overexpressing Pitx2 as a cDNA (not regulated by miR-200a) to activate the dental epithelial GRN, including amelogenin and using miR-200a overexpression to establish the epithelial cell program.
Studies based in our laboratory with the current findings establish a new method to reprogram both oral epithelial and dental mesenchymal cells to a dental epithelial cell fate (Fig. 11). Because functional dental epithelial cell cultures are a limiting resource in tissue engineering, this technique should have great potential to help produce epithelial tissues for tooth and craniofacial repair and regeneration. Human oral epithelium can be obtained from patients and reprogrammed to yield sufficient amounts of dental epithelium for tooth replacement. We are currently using these findings to initiate tooth regeneration.
FIGURE 11.
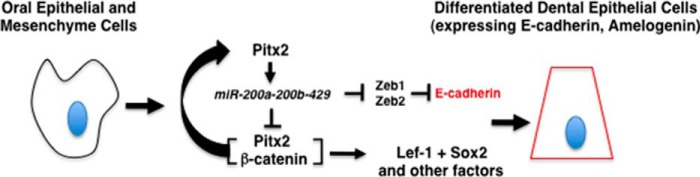
Model for the role of Pitx2 and miR-200a in cell conversion. To convert oral epithelial or dental mesenchymal cells to differentiated dental epithelium, cells are transfected with Pitx2, which regulates endogenous miR-200a expression. It has been shown that miR-200a inhibits Zeb1 and Zeb2, which repress E-cadherin expression. miR-200a feeds back to also repress Pitx2 and β-catenin expression. Pitx2 activates its own expression in concert with β-catenin providing a constant feed back loop to fine tune both Pitx2 and miR-200a expression. However, overexpression of Pitx2 cDNA drives Lef-1 and other dental epithelial factors and promotes dental epithelial cell conversion with miR-200a overexpression.
Acknowledgments
We thank the University of Iowa College of Dentistry for support and preclinical studies, the University of Iowa Carver College of Medicine Bioinformatics and RNA sequencing core. We thank Steven Eliason (University of Iowa, Carver College of Medicine) and Dr. Liu Hong (University of Iowa, College of Dentistry) for technical support.
This work was supported by University of Iowa internal funding and National Institutes of Health Grant DE13941 (to B. A. A.).
- Bmp
- bone morphogenic protein
- LaCL
- labial incisor cervical loop
- EMT
- epithelial-mesenchymal transition
- miR
- microRNA
- GRN
- gene regulatory network
- P0
- postnatal day 0.
REFERENCES
- 1. Harada H., Kettunen P., Jung H. S., Mustonen T., Wang Y. A., Thesleff I. (1999) Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J. Cell Biol. 147, 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tummers M., Thesleff I. (2003) Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development 130, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 3. Tummers M., Thesleff I. (2008) Observations on continuously growing roots of the sloth and the K14-Eda transgenic mice indicate that epithelial stem cells can give rise to both the ameloblast and root epithelium cell lineage creating distinct tooth patterns. Evol. Dev. 10, 187–195 [DOI] [PubMed] [Google Scholar]
- 4. Wang X. P., Suomalainen M., Felszeghy S., Zelarayan L. C., Alonso M. T., Plikus M. V., Maas R. L., Chuong C. M., Schimmang T., Thesleff I. (2007) An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 5, e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neubüser A., Peters H., Balling R., Martin G. R. (1997) Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell 90, 247–255 [DOI] [PubMed] [Google Scholar]
- 6. St Amand T. R., Zhang Y., Semina E. V., Zhao X., Hu Y., Nguyen L., Murray J. C., Chen Y. (2000) Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev. Biol. 217, 323–332 [DOI] [PubMed] [Google Scholar]
- 7. Lin C. R., Kioussi C., O'Connell S., Briata P., Szeto D., Liu F., Izpisúa-Belmonte J. C., Rosenfeld M. G. (1999) Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279–282 [DOI] [PubMed] [Google Scholar]
- 8. Logan C. Y., Nusse R. (2004) The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 9. Vadlamudi U., Espinoza H. M., Ganga M., Martin D. M., Liu X., Engelhardt J. F., Amendt B. A. (2005) PITX2, β-catenin, and LEF-1 interact to synergistically regulate the LEF-1 promoter. J. Cell Sci. 118, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 10. Amen M., Liu X., Vadlamudi U., Elizondo G., Diamond E., Engelhardt J. F., Amendt B. A. (2007) PITX2 and β-catenin interactions regulate Lef-1 isoform expression. Mol. Cell Biol. 27, 7560–7573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cha K. B., Douglas K. R., Potok M. A., Liang H., Jones S. N., Camper S. A. (2004) WNT5A signaling affects pituitary gland shape. Mech. Dev. 121, 183–194 [DOI] [PubMed] [Google Scholar]
- 12. Gat U., DasGupta R., Degenstein L., Fuchs E. (1998) De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell 95, 605–614 [DOI] [PubMed] [Google Scholar]
- 13. Hogan B. L. (1999) Morphogenesis. Cell 96, 225–233 [DOI] [PubMed] [Google Scholar]
- 14. Liu F., Chu E. Y., Watt B., Zhang Y., Gallant N. M., Andl T., Yang S. H., Lu M. M., Piccolo S., Schmidt-Ullrich R., Taketo M. M., Morrisey E. E., Atit R., Dlugosz A. A., Millar S. E. (2008) Wnt/β-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 313, 210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Noramly S., Freeman A., Morgan B. A. (1999) β-Catenin signaling can initiate feather bud development. Development 126, 3509–3521 [DOI] [PubMed] [Google Scholar]
- 16. Thesleff I., Nieminen P. (1996a) Tooth morphogenesis and cell differentiation. Curr. Opin. Cell Biol. 8, 844–850 [DOI] [PubMed] [Google Scholar]
- 17. Järvinen E., Salazar-Ciudad I., Birchmeier W., Taketo M. M., Jernvall J., Thesleff I. (2006) Continuous tooth generation in mouse is induced by activated epithelial Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. U.S.A. 103, 18627–18632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kratochwil K., Dull M., Farinas I., Galceran J., Grosschedl R. (1996) Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev. 10, 1382–1394 [DOI] [PubMed] [Google Scholar]
- 19. Sasaki T., Ito Y., Xu X., Han J., Bringas P., Jr., Maeda T., Slavkin H. C., Grosschedl R., Chai Y. (2005) LEF1 is a critical epithelial survival factor during tooth morphogenesis. Dev. Biol. 278, 130–143 [DOI] [PubMed] [Google Scholar]
- 20. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 21. Cao H., Wang J., Li X., Florez S., Huang Z., Venugopalan S. R., Elangovan S., Skobe Z., Margolis H. C., Martin J. F., Amendt B. A. (2010) MicroRNAs play a critical role in tooth development. J. Dent. Res. 89, 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Michon F., Tummers M., Kyyrönen M., Frilander M. J., Thesleff I. (2010) Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev. Biol. 340, 355–368 [DOI] [PubMed] [Google Scholar]
- 23. Mongroo P. S., Rustgi A. K. (2010) The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 10, 219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brabletz S., Bajdak K., Meidhof S., Burk U., Niedermann G., Firat E., Wellner U., Dimmler A., Faller G., Schubert J., Brabletz T. (2011) The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 30, 770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wellner U., Schubert J., Burk U. C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., Brunton V. G., Morton J., Sansom O., Schüler J., Stemmler M. P., Herzberger C., Hopt U., Keck T., Brabletz S., Brabletz T. (2009) The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11, 1487–1495 [DOI] [PubMed] [Google Scholar]
- 26. Korpal M., Lee E. S., Hu G., Kang Y. (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 28. Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. (2008) A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Reports 9, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brabletz S., Brabletz T. (2010) The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 11, 670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jheon A. H., Li C. Y., Wen T., Michon F., Klein O. D. (2011) Expression of MicroRNAs in the Stem Cell Niche of the Adult Mouse Incisor. PLoS One 6, e24536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cao H., Jheon A., Li X., Sun Z., Wang J., Florez S., Zhang Z., McManus M. T., Klein O. D., Amendt B. A. (2013) The Pitx2:miR-200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development 140, 3348–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morotomi T., Kawano S., Toyono T., Kitamura C., Terashita M., Uchida T., Toyoshima K., Harada H. (2005) In vitro differentiation of dental epitehlial progenitor cells through epithelial-mesenchymal interactions. Arch. Oral Biol. 50, 695–705 [DOI] [PubMed] [Google Scholar]
- 33. Tucker A., Sharpe P. (2004) The cutting-edge of mammalian development; how the embryo makes teeth. Nat. Rev. Genet. 5, 499–508 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y. D., Chen Z., Song Y. Q., Liu C., Chen Y. (2005) Making a tooth: growth factors, transcription factors and stem cells. Cell Res. 15, 301–316 [DOI] [PubMed] [Google Scholar]
- 35. Ikeda E., Morita R., Nakao K., Ishida K., Nakamura T., Takano-Yamamoto T., Ogawa M., Mizuno M., Kasugai S., Tsuji T. (2009) Fully functional bioengineered tooth replacement as an organ replacement therapy. Proc. Natl. Acad. Sci. U.S.A. 106, 13475–13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sharpe P. T., Young C. S. (2005) Test-tube teeth. Sci. Am. 293, 34–41 [DOI] [PubMed] [Google Scholar]
- 37. Yen A. H., Sharpe P. T. (2006) Regeneration of teeth using stem cell-based tissue engineering. Expert. Opin. Biol. Ther. 6, 9–16 [DOI] [PubMed] [Google Scholar]
- 38. Shi S., Bartold P. M., Miura M., Seo B. M., Robey P. G., Gronthos S. (2005) The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod. Craniofac. Res. 8, 191–199 [DOI] [PubMed] [Google Scholar]
- 39. Menicanin D., Mrozik K. M., Wada N., Marino V., Shi S., Bartold P. M., Gronthos S. (2014) Periodontal-ligament-derived stem cells exhibit the capacity for long-term survival, self-renewal, and regeneration of multiple tissue types in vivo. Stem Cells Dev. 23, 1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Menicanin D., Bartold P. M., Zannettino A. C., Gronthos S. (2010) Identification of a common gene expression signature associated with immature clonal mesenchymal cell populations derived from bone marrow and dental tissues. Stem Cells Dev. 19, 1501–1510 [DOI] [PubMed] [Google Scholar]
- 41. Gronthos S., Arthur A., Bartold P. M., Shi S. (2011) A method to isolate and culture expand human dental pulp stem cells. Methods Mol. Biol. 698, 107–121 [DOI] [PubMed] [Google Scholar]
- 42. Hynes K., Menicanin D., Mrozik K., Gronthos S., Bartold P. M. (2014) Generation of functional mesenchymal stem cells from different induced pluripotent stem cell lines. Stem Cells Dev. 23, 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 44. Ivey K. N., Srivastava D. (2010) MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 7, 36–41 [DOI] [PubMed] [Google Scholar]
- 45. Martinez N. J., Gregory R. I. (2010) MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell 7, 31–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Amendt B. A., Sutherland L. B., Russo A. F. (1999) Multifunctional role of the Pitx2 homeodomain protein C-terminal tail. Mol. Cell Biol. 19, 7001–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cox C. J., Espinoza H. M., McWilliams B., Chappell K., Morton L., Hjalt T. A., Semina E. V., Amendt B. A. (2002) Differential regulation of gene expression by PITX2 isoforms. J. Biol. Chem. 277, 25001–25010 [DOI] [PubMed] [Google Scholar]
- 48. Amendt B. A., Sutherland L. B., Semina E. V., Russo A. F. (1998) The molecular basis of Rieger syndrome: analysis of Pitx2 homeodomain protein activities. J. Biol. Chem. 273, 20066–20072 [DOI] [PubMed] [Google Scholar]
- 49. Filali M., Cheng N., Abbott D., Leontiev V., Engelhardt J. F. (2002) Wnt-3A/β-catenin signaling induces transcription from the LEF-1 promoter. J. Biol. Chem. 277, 33398–33410 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Z., Wlodarczyk B. J., Niederreither K., Venugopalan S., Florez S., Finnell R. H., Amendt B. A. (2011) Fuz regulates craniofacial development through tissue specific responses to signaling factors. PLoS One 6, e24608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cao H., Florez S., Amen M., Huynh T., Skobe Z., Baldini A., Amendt B. A. (2010) Tbx1 regulates progenitor cell proliferation in the dental epithelium by modulating Pitx2 activation of p21. Dev. Biol. 347, 289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Amen M., Espinoza H. M., Cox C., Liang X., Wang J., Link T. M., Brennan R. G., Martin J. F., Amendt B. A. (2008) Chromatin-associated HMG-17 is a major regulator of homeodomain transcription factor activity modulated by Wnt/β-catenin signaling. Nucleic Acids Res. 36, 462–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cui X. M., Shiomi N., Chen J., Saito T., Yamamoto T., Ito Y., Bringas P., Chai Y., Shuler C. F. (2005) Overexpression of Smad2 in Tgf-b3-null mutant mice rescues cleft palate. Dev. Biol. 278, 193–202 [DOI] [PubMed] [Google Scholar]
- 54. Venugopalan S. R., Amen M. A., Wang J., Wong L., Cavender A. C., D'Souza R. N., Akerlund M., Brody S. L., Hjalt T. A., Amendt B. A. (2008) Novel expression and transcriptional regulation of FoxJ1 during oro-facial morphogenesis. Hum. Mol. Genet. 17, 3643–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chavez M. G., Yu W., Biehs B., Harada H., Snead M. L., Lee J. S., Desai T. A., Klein O. D. (2013) Characterization of dental epithelial stem cells from the mouse incisor with two-dimensional and three-dimensional platforms. Tissue Eng. Part C Methods 19, 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chavez M. G., Hu J., Seidel K., Li C., Jheon A., Naveau A., Horst O., Klein O. D. (2014) Isolation and culture of dental epithelial stem cells from the adult mouse incisor. J. Vis. Exp. 10.3791/51266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuang-Hsien Hu J., Mushegyan V., Klein O. D. (2014) On the cutting edge of organ renewal: Identification, regulation, and evolution of incisor stem cells. Genesis 52, 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Z., Florez S., Gutierrez-Hartmann A., Martin J. F., Amendt B. A. (2010) MicroRNAs regulate pituitary development, and microRNA 26b specifically targets lymphoid enhancer factor 1 (Lef-1), which modulates pituitary transcription factor 1 (Pit-1) expression. J. Biol. Chem. 285, 34718–34728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen L. S., Couwenhoven R. I., Hsu D., Luo W., Snead M. L. (1992) Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ. Arch. Oral Biol. 37, 771–778 [DOI] [PubMed] [Google Scholar]
- 60. Hanks C. T., Fang D., Sun Z., Edwards C. A., Butler W. T. (1998) Dentin-specific proteins in MDPC-23 cell line. Eur. J. Oral Sci. 106, 260–266 [DOI] [PubMed] [Google Scholar]
- 61. Goecks J., Nekrutenko A., Taylor J., and Galaxy Team (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Blankenberg D., Von Kuster G., Coraor N., Ananda G., Lazarus R., Mangan M., Nekrutenko A., Taylor J. (2010) “Galaxy: a web-based genome analysis tool for experimentalists.” Curr. Protoc. Mol. Biol. 10.1002/0471142727.mb1910s89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Miller W., Kent W. J., Nekrutenko A. (2005) “Galaxy: a platform for interactive large-scale genome analysis.”. Genome Res. 15, 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Saldanha A. J. (2004) Java Treeview: extensible visualization of microarray data. Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 66. de Hoon M. J., Imoto S., Nolan J., Miyano S. (2004) Open source clustering software. Bioinformatics 20, 1453–1454 [DOI] [PubMed] [Google Scholar]
- 67. Mucchielli M. L., Mitsiadis T. A., Raffo S., Brunet J. F., Proust J. P., Goridis C. (1997) Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev. Biol. 189, 275–284 [DOI] [PubMed] [Google Scholar]
- 68. Hjalt T. A., Semina E. V., Amendt B. A., Murray J. C. (2000) The Pitx2 protein in mouse development. Dev. Dyn. 218, 195–200 [DOI] [PubMed] [Google Scholar]
- 69. Xia H., Cheung W. K., Sze J., Lu G., Jiang S., Yao H., Bian X. W., Poon W. S., Kung H. F., Lin M. C. (2010) miR-200a regulates epithelial-mesenchymal to stem-like transition via ZEB2 and β-catenin signaling. J. Biol. Chem. 285, 36995–37004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stockinger A., Eger A., Wolf J., Beug H., Foisner R. (2001) E-cadherin regulates cell growth by modulating proliferation-dependent β-catenin transcriptional activity. J. Cell Biol. 154, 1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen S., Lewallen M., Xie T. (2013) Adhesion in the stem cell niche: biological roles and regulation. Development 140, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li C. Y., Cha W., Luder H. U., Charles R. P., McMahon M., Mitsiadis T. A., Klein O. D. (2012) E-cadherin regulates the behavior and fate of epithelial stem cells and their progeny in the mouse incisor. Dev. Biol. 366, 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Karpowicz P., Willaime-Morawek S., Balenci L., DeVeale B., Inoue T., van der Kooy D. (2009) E-Cadherin regulates neural stem cell self-renewal. J. Neurosci. 29, 3885–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. DenBesten P. K., Machule D., Zhang Y., Yan Q., Li W. (2005) Characterization of human primary enamel organ epithelial cells in vitro. Arch. Oral Biol. 50, 689–694 [DOI] [PubMed] [Google Scholar]
- 75. Nakahori Y., Takenaka O., Nakagome Y. (1991) A human X-Y homologous region encodes “amelogenin.” Genomics 9, 264–269 [DOI] [PubMed] [Google Scholar]
- 76. Snead M. L., Lau E. C., Zeichner-David M., Fincham A. G., Woo S. L., Slavkin H. C. (1985) DNA sequence for cloned cDNA for murine amelogenin reveal the amino acid sequence for enamel-specific protein. Biochem. Biophys. Res. Commun. 129, 812–818 [DOI] [PubMed] [Google Scholar]
- 77. Peters H., Balling R. (1999) Teeth, where and how to make them,. TIG 15, 59–65 [DOI] [PubMed] [Google Scholar]
- 78. Jernvall J., Thesleff I. (2000) Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 92, 19–29 [DOI] [PubMed] [Google Scholar]
- 79. Zeichner-David M., Diekwisch T., Fincham A., Lau E., MacDougall M., Moradian-Oldak J., Simmer J., Snead M., Slavkin H. C. (1995) Control of ameloblast differentiation. Int. J. Dev. Biol. 39, 69–92 [PubMed] [Google Scholar]
- 80. Mustonen T., Tümmers M., Mikami T., Itoh N., Zhang N., Gridley T., Thesleff I. (2002) Lunatic fringe, FGF, and BMP regulate the Notch pathway during epithelial morphogenesis of teeth. Dev. Biol. 248, 281–293 [DOI] [PubMed] [Google Scholar]
- 81. Limeback H. (1987) Enamel protein and collagen production by cells subcultured from porcine tooth bud explants. Biochem. Cell Biol. 65, 698–709 [DOI] [PubMed] [Google Scholar]
- 82. Farges J. C., Couble M. L., Joffre A., Hartmann D. J., Magloire H. (1991) Morphological and immunocytochemical characterization of cultured rat incisor cervical epithelial cells. Arch. Oral Biol. 36, 737–745 [DOI] [PubMed] [Google Scholar]
- 83. Tabata M. J., Matsumura T., Liu J. G., Wakisaka S., Kurisu K. (1996) Expression of cytokeratin 14 in ameloblast-lineage cells of the developing tooth of rat, both in vivo and in vitro. Arch. Oral Biol. 41, 1019–1027 [DOI] [PubMed] [Google Scholar]
- 84. Matsumura T., Tabata M. J., Wakisaka S., Sakuda M., Kurisu K. (1998) Ameloblast-lineage cells of rat tooth germs proliferate and scatter in response to hepatocyte growth factor in culture. Int. J. Dev. Biol. 42, 1137–1142 [PubMed] [Google Scholar]
- 85. Tabata M. J., Matsumura T., Fujii T., Abe M., Kurisu K. (2003) Fibronectin accelerates the growth and differentiation of ameloblast lineage cells in vitro. J. Histochem. Cytochem. 51, 1673–1679 [DOI] [PubMed] [Google Scholar]
- 86. Kawano S., Saito M., Handa K., Morotomi T., Toyono T., Seta Y., Nakamura N., Uchida T., Toyoshima K., Ohishi M., Harada H. (2004) Characterization of dental epithelial progenitor cells derived from cervical-loop epithelium in a rat lower incisor. J. Dent. Res. 83, 129–133 [DOI] [PubMed] [Google Scholar]
- 87. Harada H., Toyono T., Toyoshima K., Yamasaki M., Itoh N., Kato S., Sekine K., Ohuchi H. (2002) FGF10 maintains stem cell compartment in developing mouse incisors. Development 129, 1533–1541 [DOI] [PubMed] [Google Scholar]
- 88. Harada H., Ohshima H. (2004) New perspectives on tooth development and the dental stem cell niche. Arch. Histol. Cytol. 67, 1–11 [DOI] [PubMed] [Google Scholar]
- 89. Maas R., Bei M. (1997) The genetic control of early tooth development. Crit. Rev. Oral. Biol. Med. 8, 4–39 [DOI] [PubMed] [Google Scholar]
- 90. Thesleff I., Tummers M. (2009) Tooth organogenesis and regeneration. The Stem Cell Research Community, StemBook [PubMed] [Google Scholar]
- 91. Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., Lee-Messer C., Dolmetsch R. E., Tsien R. W., Crabtree G. R. (2011) MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gay I., Cavender A., Peto D., Sun Z., Speer A., Cao H., Amendt B. A. (2014) Differentiation of human dental stem cells reveals a role for microRNA-218. J. Periodontal Res. 49, 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cui L., Guan Y., Qu Z., Zhang J., Liao B., Ma B., Qian J., Li D., Li W., Xu G. T., Jin Y. (2013) WNT signaling determine tumorigenicity and function of ESC-derived retinal progenitors. J. Clin. Invest. 123, 1647–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liu F., Millar S. E. (2010) Wnt/β-catenin signaling in oral tissue development and disease. J. Dent. Res. 89, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bei M. (2009) Molecular genetics of tooth development. Curr. Opin. Genet. Dev. 19, 504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liu W., Selever J., Lu M. F., Martin J. F. (2003) Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development 130, 6375–6385 [DOI] [PubMed] [Google Scholar]