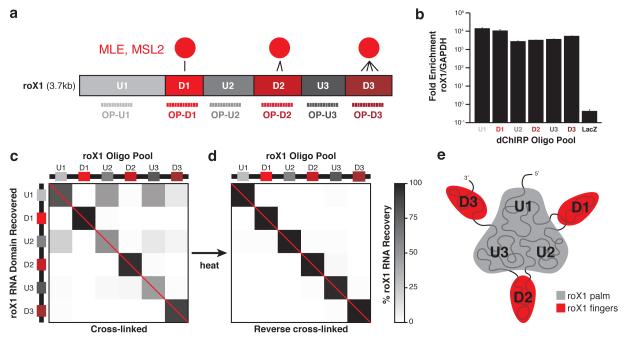
Figure 2.
dChIRP RNA co-recovery reveals roX1’s topological architecture. (a) Schematic representation of known roX1 domain interactions with MLE protein and dChIRP OP design strategy. MLE directly contacts the three D domains (D1, D2, and D3). The three intervening U domains (U1, U2, and U3) exhibit minimal binding. Six OPs were designed to target and recover each domain. (b) roX1 dChIRP specifically enriches for roX1 RNA. roX1 RNA is >1000-fold enriched over the abundant GAPDH mRNA in roX1 dChIRP samples. LacZ ChIRP does not enrich for roX1 over GAPDH. Average of technical triplicates +s.d. shown. (c, d) roX1 RNA recovery by dChIRP. To confirm that roX1 dChIRP successfully recovers the targeted RNA domain, the RNA fraction of each dChIRP sample was analyzed by RT-qPCR, using primers within each domain of roX1. Within each sample, roX1 domain recovery was quantified against input and normalized to total roX1 RNA recovery (% roX1 RNA recovery). As expected, each OP best enriches for the target roX1 domain (b, red diagonal). Whereas domains D1, D2, and D3 were recovered independently, domains U1, U2, and U3 were co-recovered. To demonstrate that the co-recovery of domains U1, U2, and U3 is cross-linking-dependent, fixed chromatin was thermally de-cross-linked and dChIRP was performed (c). Each of the domains of roX1 is independently recovered. (e) Schematic representation of roX1 intramolecular topology. Domains U1, U2, and U3 are topologically proximal to one another, forming the core “palm” of roX1. Domains D1, D2, and D3 extend as “fingers” and are distant from one another and the intervening U domains.