Abstract
Objective
Validation of a new fast software technique to segment the cartilage on knee MR acquisitions. Large studies of knee osteoarthritis (OA) will require fast and reproducible methods to quantify cartilage changes for knee MR data. In this report we document and measure the reproducibility and reader time of a software-based technique to quantify the volume and thickness of articular cartilage on knee MR images.
Methods
The software was tested on a set of duplicate sagittal 3D DESS acquisitions from 15 (8 OA, 7 normal) subjects. The repositioning, inter-reader, and intra-reader reproducibility of the cartilage volume and thickness were measured independently as well as the reader time for each cartilage plate. The root-mean square coefficient of variation (RMSCoV) was used as metric to quantify the reproducibility of cartilage volume (VC) and mean cartilage thickness (ThC).
Results
The repositioning RMSCoV was VC = 2.0% and ThC = 1.2% (femur), VC = 2.9% and ThC = 1.6% (medial tibial plateau), VC = 5.5% and ThC = 2.4% (lateral tibial plateau), VC = 4.6 % and ThC = 2.3% (patella). RMSCoV values were higher for the inter-reader reproducibility (VC: 2.5 % - 8.6 %) (ThC: 1.9% - 5.2%) and lower for the intra-reader reproducibility (VC: 1.6 % - 2.5 %) (ThC: 1.2% - 1.9%). The method required an average of 75.4 minutes per knee.
Conclusions
We have documented a fast reproducible semi-automated software method to segment articular cartilage on knee MR acquisitions.
Keywords: Cartilage, osteoarthritis, magnetic resonance imaging, segmentation, software
I. INTRODUCTION
Osteoarthritis (OA) of the knee is a common disease of middle age and older adults associated with disability and very high economic and social costs. Radiological imaging offers quantifiable outcome measures for knee OA, which can be used in studies to evaluate therapies(1-3). Magnetic resonance (MR) imaging, in particular, is a powerful tool since it provides visualization of the interarticular cartilage in three dimensions (3D). Knee MR can be assessed for OA using semi-quantitative scoring systems (4, 5), however image analysis software techniques offer direct measurement of the size and shape of articular cartilage.
Image-processing software is used for computer-aided diagnosis of disease with most radiological modalities and for numerous clinical applications. For 3D knee MR images, the goal of such software is to segment the cartilage from the surrounding tissue so that the volume and thickness can be calculated. There are several different software tools published in the literature and significant effort continues by several laboratories in this area(6-16).
The development of a fully automated software tool to segment the cartilage on knee MR images could be considered the ultimate goal of these endeavors. Such software would perform the segmentation without any intervention from an individual and the reader time would be essentially zero. Fully automated segmentation, however, will be very difficult if not impossible due to ambiguous cartilage margins in many areas of the images.
Severely diseased knees, in particular, offer a much greater challenge and present subtleties along the cartilage interface in the image that require expertise and training to interpret. To date there has been no fully automated software technique reported in the literature.
Rather than attempt to develop fully automated software, we have concentrated our efforts on a hybrid approach. The segmentation tool requires an expert reader with anatomic knowledge to guide the software, but the user is provided with numerous automated image-processing tools to increase the speed of cartilage segmentation. Validation consists of measuring the precision of the segmentation method as well as the reader time.
Our study used data from a pilot study for the Osteoarthritis Initiative. The Osteoarthritis Initiative (OAI) is a program jointly sponsored by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), several other Institutes at the National Institutes of Health (NIH), and the pharmaceutical industry. It is targeted at identifying the most potent OA biomarkers for analyzing development and progression of symptomatic knee osteoarthritis. The OAI has an enrollment of approximately 4,500 subjects(17) and will acquire MR exams on each subject at baseline and during four annual follow-up visits. In addition to the OAI, there are several very large OA studies that include knee MR in their protocols(18, 19). The goal of this study is to measure the reader and repositioning precision of the semi-automated software method using two readers and duplicate MRI acquisitions.
II. MATERIALS AND METHODS
II.A Data Set
As part of a pilot study for the Osteoarthritis Initiative, test-retest knee MR exams were performed for 19 participants with no to moderate degrees of clinical OA (9 normal and 10 OA) (20). The data were acquired using a Siemens (Erlangen, Germany) Trio 3T MR system and a USA Instruments (Aurora, OH) quadrature transmit/receive extremity coil using a 3D sagittal DESS (dual echo steady state) with water excitation. The slice thickness was 0.7 mm (160 slices per knee) and the in-plane pixel size was 0.37 mm × 0.46 mm, interpolated to 0.37 mm × 0.37 mm. Each subject was scanned, removed from the magnet, walked for 10 minutes, and then was rescanned on the same visit so that the repositioning reproducibility could be measured; this provided a total of 38 MR acquisitions for segmentation.
The MR exams from four randomly chosen subjects (two normal and two OA) served as a training set for the readers and to optimize the software. The remaining 15 subjects were randomly divided into two groups, Set 1 and Set 2, consisting of 8 and 7 subjects respectively, and the study employed two readers, Reader 1 (G.N.) and Reader 2 (M.B.) Reader 1 used the software tool to segment the data in Set 1 (16 acquisitions), while Reader 2 read Set 2 (14 acquisitions). To measure the inter-reader reproducibility, Reader 1 also read a single acquisition, of the pair, for the subjects in Set 2 (7 acquisitions), and Reader 2 similarly read a single acquisition from Set 1 (8 acquisitions). For measuring the intra-reader reproducibility the readers were asked to reread a single exam for each patient from the originally assigned set. To reduce memory effect, the second reading took place more than one month after the last previous reading. The data were randomized as to time order and patient ID, and the readers were fully blinded.
II.B Description of software tool
The software program consists of two parts: the low-level image-processing algorithms and the graphical user interface (GUI) tool. The core image-processing code was written using the C programming language and consisted of customized edge-tracking algorithms and an active-contour edge refinement procedure. Modification to the edge detection routines were developed for specific regions of the knee cartilage. As an example, the cartilage-soft tissue interface presents a different edge-detection challenge than the cartilage-bone margin; the software tool employed different algorithms for these two locations. The higher-level GUI tool is the link between the reader and the low-level procedures and was written with the Interactive Data Language (IDL), (ITT Visual Information Solutions, Boulder, CO). The GUI also provides numerous semi-automated editing functions that allow the reader to correct software mistakes. The software runs on a standard personal computer using the Windows operating system. Figure 1 shows a screen capture of a computer running the application.
Figure 1.
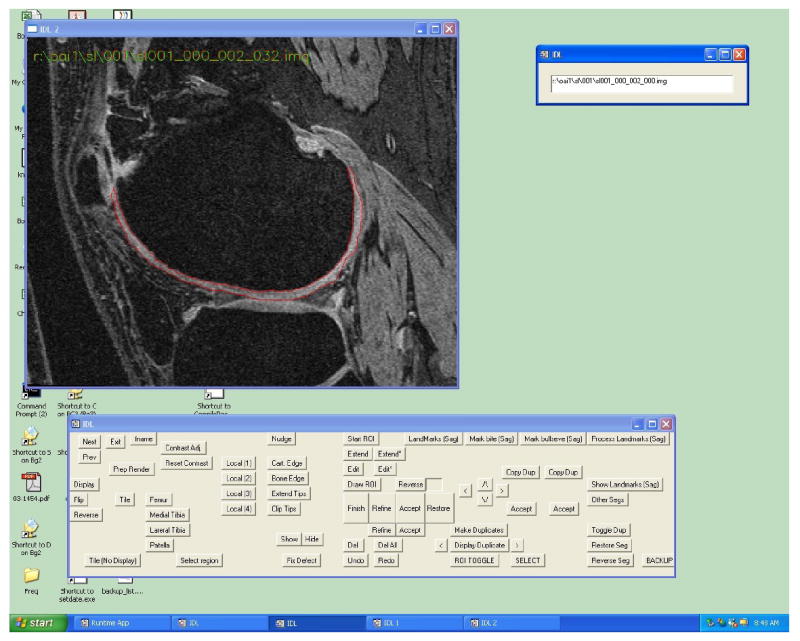
Screen capture of the software tool.
The method functions by performing a two-dimensional segmentation on each slice of the MR acquisition. The reader first selects a slice near the center of the cartilage plate and places a seed point on the bone-cartilage margin. The software then employs an automated edge-tracking algorithm to attempt a segmentation of the cartilage on this slice. (Figure 2a) Since extraneous edges often cause the software to deviate from the true margin, additional editing tools can be employed to guide the segmentation. The initial slice typically requires less than 30 seconds of reader time for segmentation. Figure 2b shows the segmented slice. The software then initiates an automated active-contour algorithm to refine the segmentation. (Figure 2c)
Figure 2.
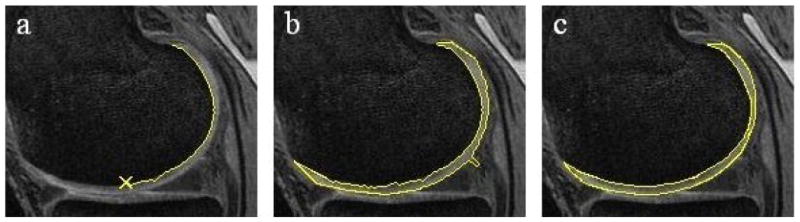
Example of the segmentation procedure on the central slice.
Once a central slice is segmented, the software proceeds to an adjacent slice using the computer-delineated margins from the previous slice and an active contour edge detection algorithm to attempt an automated segmentation. In regions where the software segmentation fails, there are user tools for convenient editing of the computer-determined contours. This process continues on a slice-by-slice basis until the reader judges that the end of the cartilage plate has been reached. The second half of the cartilage plate is then segmented starting at the central slice and proceeding in the opposite direction.
II.C Segmentation Study
The readers were instructed to use the software tool to segment the total femur, medial tibia, lateral tibia and patella cartilage. The volume and average thickness were calculated for each plate. A comparison of the duplicate exams was used to establish the repositioning reproducibility while multiple readings of the same acquisition were used to measure the intra and inter-reader reproducibility. The root-mean square coefficient of variation (RMSCoV) was used as a metric to quantify the reproducibility. The readers were also asked to record the time required to segment each cartilage plate. We define the RMSCoV as:
where SD denotes the standard deviation and N is the number of pairs. SD is defined as:
where n is the number of measurements. For duplicate readings, n = 2.
III. RESULTS
Table 1 gives the results for the reproducibility values for all subjects. Tables 2 and 3 provide the results for the reproducibility values for the normal and OA subjects individually. Table 4 shows the average reader time for each plate. The repositioning reproducibility ranged from 2.0 % to 5.5 % (VC), and 1.2 % to 2.5 % (ThC). Reproducibility was lower for the normal compared to OA knees but the difference was not dramatic. Application of a one-sided t-test using the absolute difference of the percent difference between pairs showed no statistically significant difference between the OA and normal knees. The average segmentation time was 75.4 minutes per knee, with a modest increase required for the OA versus the normal knees.
Table 1.
Reproducibility values (RMSCoV) for all subjects (N = 15).
| All Subjects | ||||
|---|---|---|---|---|
| Femur | Medial tibial plateau | Lateral tibial plateau | Patella | |
| Repositioning | ||||
| Volume (VC) | 2.0 % | 2.9 % | 5.5 % | 4.6 % |
| Thickness (ThC) | 1.2 % | 1.6 % | 2.4 % | 2.3 % |
| Inter-reader | ||||
| Volume(VC) | 2.5 % | 2.8 % | 8.6 % | 3.3 % |
| Thickness (ThC) | 1.9 % | 2.5 % | 5.2 % | 3.3 % |
| Intra-reader | ||||
| Volume(VC) | 1.6 % | 3.0 % | 3.4 % | 3.5 % |
| Thickness (ThC) | 1.2 % | 2.1 % | 1.7 % | 1.9 % |
Table 2.
Reproducibility values (RMSCoV) for the normal subjects (N = 7).
| Normal Subjects | ||||
|---|---|---|---|---|
| Femur | Medial tibial plateau | Lateral tibial plateau | Patella | |
| Repositioning | ||||
| Volume(VC) | 1.5 % | 3.6 % | 4.8 % | 2.5 % |
| Thickness (ThC) | 1.0 % | 1.5 % | 1.8 % | 1.8 % |
| Inter-reader | ||||
| Volume(VC) | 1.8 % | 2.3 % | 5.6 % | 2.9 % |
| Thickness (ThC) | 1.9 % | 1.2 % | 6.5 % | 2.3 % |
| Intra-reader | ||||
| Volume(VC) | 1.0 % | 2.3 % | 3.2 % | 2.7 % |
| Thickness (ThC) | 0.5 % | 1.4 % | 1.7 % | 0.6 % |
Table 3.
Reproducibility values (RMSCoV) for the OA subjects (N = 8).
| OA Subjects | ||||
|---|---|---|---|---|
| Femur | Medial tibial plateau | lateral tibial plateau | Patella | |
| Repositioning | ||||
| Volume(VC) | 2.3 % | 1.9 % | 6.1 % | 5.9 % |
| Thickness (ThC) | 1.3 % | 1.8 % | 2.8 % | 2.6 % |
| Inter-reader | ||||
| Volume(VC) | 2.9 % | 3.2 % | 10.6 % | 3.6 % |
| Thickness (ThC) | 2.0 % | 3.2 % | 3.6 % | 4.0 % |
| Intra-reader | ||||
| Volume(VC) | 2.1 % | 3.5 % | 3.6 % | 4.1 % |
| Thickness (ThC) | 1.6 % | 2.5 % | 1.7 % | 2.5 % |
Table 4.
Average reader times in minutes.
| All subjects (N = 15) | Normal subjects (N = 7) | OA subjects (N = 8) | |
|---|---|---|---|
| Femur | 39.9 | 34.6 | 42.8 |
| Medial tibial plateau | 11.8 | 10.7 | 12.4 |
| Lateral tibial plateau | 10.2 | 9.3 | 10.4 |
| Patella | 13.5 | 11.2 | 14.9 |
| Total knee | 75.4 | 66.9 | 80.6 |
The repositioning reproducibility for the total femur and medial tibial plateau was excellent but the method was less reproducible for the lateral tibial plateau and patella. The values are similar to the results from other studies utilizing independent analysis methods. Direct comparison can be difficult since the number of subjects is small and the MR image contrast is somewhat different, however, our results compare favorably with one study that used the same data set(16).
IV DISCUSSION
A close examination of the outlier cases demonstrated some degree of reader misinterpretation and the potential for improvement of the segmentation algorithms, graphical user interface, and work flow. For the lateral tibial plateau much of the reproducibility error occurred in the anterior and posterior portions of the plate where adjacent inter-articular fluid obscured the cartilage margin. The implementation of improved reader training and a centralized quality assurance step may mitigate some of the interpretation errors. Longitudinal studies will use paired readings of the baseline and follow-up exams, which will also reduce this problem. We are implementing a paired reading procedure into the software tool.
On average our technique required 75.4 minutes to segment all four cartilage plates of the knee in the sagittal plane. Comparison to other techniques is difficult since very few studies report this variable. In McWalter et. al(13) reader times ranging from 57 to 78 minutes for a single (patella or medial tibial plateau) cartilage plate are quoted. Our method is faster by a factor of four to five times, however the McWalter study used a reader, “with no previous experience in cartilage segmentation.” Each of our readers had participated in a previous study as part of the OAI pilot project. In its current state our semi-automated segmentation tool can feasibly handle the data analysis load for large studies involving hundreds of knee MR exams without an excessive reader cost. As with the reproducibility there was a modest difference in reader time between normal and OA knees since diseased cartilage was more likely to cause algorithm failures.
Of the three reproducibility measures, the intra-reader variation was smallest while the inter-reader reproducibility was highest. Since most studies use a single reader for each cartilage plate, demonstrating good inter-reader reproducibility is less critical than intra-rater or repositioning reproducibility. The higher RMSCoV values may also indicate that our study could benefit from more systematic reader training, or a final quality assurance step performed by a single expert. As expected, the reproducibility was better for the normal subjects, although the effect of disease state was not dramatic.
We found that the (ThC) reproducibility was systematically smaller compared to (VC). Errors in segmentation may affect the VC measurement more since it is a 3D measure while ThC is fundamentally one-dimensional. Slight changes in the delineated margins could potentially cause a greater proportional change to the total volume compared to the average thickness. This is analogous to the case of the percent error introduced to the volume of a sphere by a change in the radius; the volume is proportional to the radius cubed. Determining the potential value of ThC over VC will ultimately require longitudinal data. While a measure may be more reproducible, it could also be less disease sensitive and therefore less value as a surrogate out come measure. As cartilage wears in an OA patient, the average thickness may not change substantially especially if the loss occurs in a local region. The measurement of change may be lost when averaging over the remainder of non diseased cartilage. However once the technique can measure cartilage for smaller subregions or especially for very local defect areas, the thickness measurement may be superior. Such an approach would require the implementation of 3D image registration so that the baseline and follow-up scans can be accurately compared.
An examination of the individual components comprising the RMSCoV measurement showed that they were dominated by a few “difficult” cases where segmentation was more challenging. For this reason the t-test may not be appropriate since the distribution of percent differences is unlikely to be normal. Also this effect impedes our ability to draw definitive conclusions about the performance of the method for different plates, and disease state. As an example, the elimination of two cases for the lateral tibial plateau will make the reproducibility similar to the other plates. A higher powered validation study will be necessary to better understand the more subtle details.
There are several limitations of our study. First the number of subjects, N = 15, was relatively small. We evaluated the software tool on images acquired with one type of acquisition and therefore have not demonstrated a more general applicability. In practice, we would likely make changes to the semi-automated steps to customize the tool for different MR acquisitions, spatial resolution, and noise conditions. We have designed the software architecture to facilitate customized modifications. The study evaluated reproducibility for the full cartilage plate; an examination of subregions or localized defects may also be important. Finally, the study used cross sectional data; our methods should be evaluated using longitudinal data to validate the sensitivity of the method to detect change. In this paper we report the precision of the technique; measuring accuracy is also crucial and will be the subject of a future study.
We envision several future directions for our work. Additional software development to increase the level of automation should both improve the reproducibly and decrease the reader time. We are confident that reader times of 10 - 20 minutes for the full knee are possible while also improving the reproducibility. Until such reduced times are achieved large OA studies, such as the OAI, will have excessive reader cost. We also anticipate making additions to the software for segmenting subregions and focal defects, 3D image registration, and performing an evaluation study using longitudinal data.
V. CONCLUSIONS
We have documented a reproducible and fast semi-automated software technique to segment the articular cartilage on high resolution 3D MR acquisitions of the knee. This technique has the potential for use in the analysis of MR data from moderately large studies of knee OA.
Acknowledgments
The Osteoarthritis Initiative (OAI) and this pilot study are conducted and supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) in collaboration with the OAI Investigators and Consultants. This manuscript has been reviewed by the OAI Publications committee for scientific content and data interpretation. The research reported in this article was supported in part by contracts N01-AR-2-2261, N01-AR-2-2262 and N01-AR-2-2258 from NIAMS. We are grateful to the Ohio State University team, particularly Kim Toussant, and to the Center for Primary Care and Prevention team at Memorial Hospital of Rhode Island for recruitment of the study subjects and to Larry Martin RTR(MR) and Lynn Fanella RTR(MR) for acquiring the MR images. David White, PhD is thanked for his help with the initial study setup and for randomization of the images.
This work was also supported by a contract with the NIH/NIAMS intramural program. We would like to thank Raphaela Goldbach-Mansky of the NIH/NIAMS Intramural Research Program for her support in developing early versions of the software.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gray ML, Eckstein F, Peterfy C, Dahlberg L, Kim YJ, Sorensen AG. Toward imaging biomarkers for osteoarthritis. Clin Orthop Relat Res. 2004:S175–181. doi: 10.1097/01.blo.0000144972.50849.d9. [DOI] [PubMed] [Google Scholar]
- 2.Eckstein F, Ateshian G, Burgkart R, Burstein D, Cicuttini F, Dardzinski B, et al. Proposal for a nomenclature for Magnetic Resonance Imaging based measures of articular cartilage in osteoarthritis. Osteoarthritis Cartilage. 2006;14:974–983. doi: 10.1016/j.joca.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Bauer DC, Hunter DJ, Abramson SB, Attur M, Corr M, Felson D, et al. Classification of osteoarthritis biomarkers: a proposed approach. Osteoarthritis Cartilage. 2006:723–727. doi: 10.1016/j.joca.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Biswal S, Hastie T, Andriacchi TP, Bergman GA, Dillingham MF, Lang P. Risk factors for progressive cartilage loss in the knee: a longitudinal magnetic resonance imaging study in forty-three patients. Arthritis Rheum. 2002;46:2884–2892. doi: 10.1002/art.10573. [DOI] [PubMed] [Google Scholar]
- 5.Peterfy CG, Guermazi A, Zaim S, Tirman PF, Miaux Y, White D, et al. Whole-Organ Magnetic Resonance Imaging Score (WORMS) of the knee in osteoarthritis. Osteoarthritis Cartilage. 2004;12:177–190. doi: 10.1016/j.joca.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Peterfy CG, van Dijke CF, Janzen DL, Gluer CC, Namba R, Majumdar S, et al. Quantification of articular cartilage in the knee with pulsed saturation transfer subtraction and fat-suppressed MR imaging: optimization and validation. Radiology. 1994;192:485–491. doi: 10.1148/radiology.192.2.8029420. [DOI] [PubMed] [Google Scholar]
- 7.Solloway S, Hutchinson CE, Waterton JC, Taylor CJ. The use of active shape models for making thickness measurements of articular cartilage from MR images. Magn Reson Med. 1997;37:943–952. doi: 10.1002/mrm.1910370620. [DOI] [PubMed] [Google Scholar]
- 8.Kshirsagar AA, Watson PJ, Tyler JA, Hall LD. Measurement of localized cartilage volume and thickness of human knee joints by computer analysis of three-dimensional magnetic resonance images. Invest Radiol. 1998;33:289–299. doi: 10.1097/00004424-199805000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Stammberger T, Eckstein F, Michaelis M, Englmeier KH, Reiser M. Interobserver reproducibility of quantitative cartilage measurements: comparison of B-spline snakes and manual segmentation. Magn Reson Imaging. 1999;17:1033–1042. doi: 10.1016/s0730-725x(99)00040-5. [DOI] [PubMed] [Google Scholar]
- 10.Lynch JA, Zaim S, Zhao J, Stork A, Peterfy C, Genant HK. Cartilage Segmentaiton of 3D MRO scans of the osteoartiritic knee combining user knowlege and active contours. Proc SPIE. 2000;3979:925–935. [Google Scholar]
- 11.Cicuttini F, Forbes A, Asbeutah A, Morris K, Stuckey S. Comparison and reproducibility of fast and conventional spoiled gradient-echo magnetic resonance sequences in the determination of knee cartilage volume. J Orthop Res. 2000;18:580–584. doi: 10.1002/jor.1100180410. [DOI] [PubMed] [Google Scholar]
- 12.Steines D, Cheng C, Wong J, Tsai S, Napel P, Lang P. Segmentation of Osteoarthritic Femoral Cartilage Using Live Wire. Proc Intl Soc Mag Reson Med. 2000;8:220. [Google Scholar]
- 13.McWalter EJ, Wirth W, Siebert M, von Eisenhart-Rothe RM, Hudelmaier M, Wilson DR, et al. Use of novel interactive input devices for segmentation of articular cartilage from magnetic resonance images. Osteoarthritis Cartilage. 2005;13:48–53. doi: 10.1016/j.joca.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Tamez-Pena J, Barbu-McInnis M, Totterman S. Unsupervised definition of the tibia-femoral joint regions of the human knee and its applications to cartilage analysis. Proceedings of SPIE. 2006;6144:1465–1475. [Google Scholar]
- 15.Raynauld JP, Martel-Pelletier J, Berthiaume MJ, Beaudoin G, Choquette D, Haraoui B, et al. Long term evaluation of disease progression through the quantitative magnetic resonance imaging of symptomatic knee osteoarthritis patients: correlation with clinical symptoms and radiographic changes. Arthritis Res Ther. 2006;8:R21. doi: 10.1186/ar1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckstein F, Hudelmaier M, Wirth W, Kiefer B, Jackson R, Yu J, et al. Double echo steady state magnetic resonance imaging of knee articular cartilage at 3 Tesla: a pilot study for the Osteoarthritis Initiative. Ann Rheum Dis. 2006;65:433–441. doi: 10.1136/ard.2005.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osteoarthritis Initiative. 2006 http://www.niams.nih.gov/ne/oi/index.htm (checked 5/2006)
- 18.Visser M, Newman AB, Nevitt MC, Kritchevsky SB, Stamm EB, Goodpaster BH, et al. Reexamining the sarcopenia hypothesis. Muscle mass versus muscle strength. Health, Aging, and Body Composition Study Research Group. Ann N Y Acad Sci. 2000;904:456–461. [PubMed] [Google Scholar]
- 19.Multicenter Osteoarthritis Study. 2006 http://researchresources.bumc.bu.edu/abstract/5U01AG018820-05.htm (checked 5/2006)
- 20.Schneider E. Pilot Study Analyses: Results and Perspectives. Presented at the 10th World Congress of the Osteoarthritis Research Society International (OARSI); Boston Mass. 2005. [Google Scholar]


