Abstract
DNA double-strand breaks are repaired by two major pathways, homologous recombination or nonhomologous end joining. The commitment to one or the other pathway proceeds via different steps of resection of the DNA ends, which is controlled and executed by a set of DNA double-strand break sensors, endo- and exonucleases, helicases, and DNA damage response factors. The molecular choreography of the underlying protein machinery is beginning to emerge. In this review, we discuss the early steps of genetic recombination and double-strand break sensing with an emphasis on structural and molecular studies.
The early steps of homologous recombination are becoming clearer. Of particular interest is the MRN complex, which recognizes double-strand breaks, performs initial resection, and sets off a DNA damage response network.
All domains of life maintain genomes and ensure genetic diversity through homologous recombination (HR) or homology directed repair. HR is initiated by single unprotected DNA ends, which arise at collapsed replication forks and unprotected telomeres, or by DNA double-strand breaks (DSBs), which are products of ionizing radiation, reactive oxygen species, genotoxic chemicals, or abortive topoisomerase reactions (Sutherland et al. 2000; Aguilera and Gomez-Gonzalez 2008; Cadet et al. 2012; Mehta and Haber 2014). In special cellular states, programmed DSBs are introduced by endonucleases to initiate the generation of genetic variability by processes such as meiotic recombination of homologous chromosomes (Lam and Keeney 2014; Zickler and Kleckner 2014), V(D)J and class switch recombination to generate antibody diversity and yeast-mating-type switching (Gapud and Sleckman 2011; Haber 2012; Xu et al. 2012b). Failure to repair DSBs can lead to cell death or gross chromosomal aberrations, which in humans are a hallmark of cancer (Myung et al. 2001a,b; Hanahan and Weinberg 2011).
Beside HR, DSBs can also be repaired by nonhomologous end joining (NHEJ). Although HR requires a template such as a sister chromatid or a homologous chromosome and is limited to S and G2 phases of the cell cycle, NHEJ is template-independent and can occur in all cell cycle states. Indeed, the choice of pathways is to a significant extent not stochastic but a function of the cell cycle (Ferretti et al. 2013), with NHEJ being the predominant pathway in mammals outside of S phase. NHEJ is basically a ligation reaction of two DNA ends that are only minimally processed. Derivatives of NHEJ such as microhomology-mediated end joining (MMEJ) or alternative NHEJ (alt-NHEJ) require more substantial processing and may lead to the loss of genetic information. For recent reviews of NHEJ, which is not covered in detail here, please refer to, for example, Thompson (2012) and Chiruvella et al. (2013).
HR has multiple steps and requires extensive processing of DNA ends (Symington 2014). First, the free DNA ends are recognized by DSB sensors, followed by 5′-3′ resection of the DNA ends. In eukaryotes and archaea, this step may be divided into initial short-range resection, after which MMEJ/alt-NHEJ can still occur, followed by processive long-range resection that commits the pathway to HR. The 3′ single-stranded DNA (ssDNA) filament, bound by the DNA strand exchange protein RecA/Rad51, pairs with the homologous sequence on the template and thus forms a D-loop. The 3′ tail serves as a primer for a repair polymerase and is extended by using the homologous strand as template, a process that “restores” the disrupted genetic information. Various pathways involve the displacement of the free strand, the capture of the second strand to form Holliday junctions, or the cleavage of the D-loop (Mehta and Haber 2014).
In this review, we focus on structural aspects of the early steps in homologous recombination. Of particular interest is the Mre11-Rad50-Nbs1 (MRN) complex, which recognizes DSBs, performs initial resection, and sets off a DNA damage response (DDR) signaling network. We further discuss the nucleases and helicases that are involved in long-range resection. Recent reviews of later steps in HR, which are not covered here, have been published elsewhere (Amunugama and Fishel 2012; Chiruvella et al. 2013; Jasin and Rothstein 2013).
DSB END RECOGNITION
The Mre11-Rad50-Nbs1 Complex
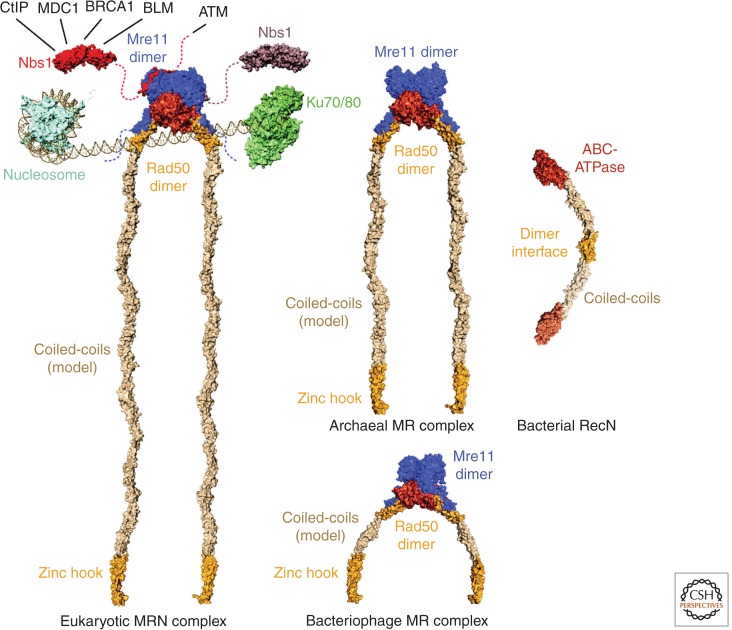
Among the early and central players in DNA end metabolism are Ku and the Mre11-Rad50-Nbs1 (MRN) complex, which are considered “sensors” for DSBs. Ku binds to DNA ends as a ring-shaped heterodimer (Fig. 1) consisting of Ku70/Ku80 and initiates NHEJ (Walker et al. 2001; Chiruvella et al. 2013). The Saccharomyces cerevisiae MRN homolog, Mre11-Rad50-Xrs2 (MRX), has been shown to be one of the first complexes that are recruited to DSBs (Lisby et al. 2004). MRN is involved in the selection of DSB repair pathways that require end resection (HR, MMEJ, alt-NHEJ) as opposed to NHEJ (Truong et al. 2013). Homologs of Mre11 and Rad50 (MR) are present in all domains of life and may be fused into a single peptide chain (Yoshida et al. 2011).
Figure 1.
The Mre11-Rad50-Nbs1 complex and phylogenetic orthologs. Structural model of MR(N) complexes together with a nucleosome, the Ku-DNA complex and RecN. Nbs1 interaction partners are indicated. The eukaryotic MRN model was built from Schizosaccharomyces pombe MN and Nbs1 (PDB code 4FBW, Schiller et al. 2012), Methanocaldococcus jannaschii MR, Pyrococcus furiosus Zn-hook and a coiled-coil model. The archaeal model is based on the M. jannaschii MR structure and the P. furiosus Zn-hook. Bacteriophage MR is modeled on the Thermotoga maritima MR complex together with the P. furiosus Zn-hook and a coiled-coil model. PDB codes are 1AOI (nucleosome, Luger et al. 1997), 1JEY (Ku-DNA complex, Walker et al. 2001), 4AD8 and 4ABX (RecN, Pellegrino et al. 2012), 4FBW (MN complex, Schiller et al. 2012), 3HUE (Nbs1, Williams et al. 2009), 3AVO (MR complex, Lim et al. 2011), and 1L8D (Zn-hook, Hopfner et al. 2002).
MRN is a multifunctional ATP-regulated nuclease with endo- and exonuclease activity and long structural tails. In vitro, the MR(N) complex is able to partially melt and unwind DNA and displays both 3′ to 5′ exonuclease and ssDNA endonuclease activity to process DSBs (Connelly et al. 1997, 1999; Furuse et al. 1998; Paull and Gellert 1998; Trujillo et al. 1998; Hopfner et al. 2000a, 2001; Trujillo and Sung 2001; Lobachev et al. 2002; Hopkins and Paull 2008; Cannon et al. 2013). Bacteriophage T4 also possesses homologs of Mre11 and Rad50 (gp46/gp47), which play an essential role in initiation of recombination-dependent replication at later stages of infection (Kreuzer and Brister 2010; Almond et al. 2013). In bacteria, MR (denoted SbcCD) degrades hairpin structures in the wake of replication forks and protects the cell against inverted chromosome duplication together with RecA (Zahra et al. 2007; Eykelenboom et al. 2008; Darmon et al. 2010). In archaea, like in eukaryotes, MR(N) is recruited to and repairs DSBs that are induced using ionizing radiation or genotoxic agents and that arise at stalled replication forks (Costanzo et al. 2001; Neale et al. 2005; Trenz et al. 2006; Frols et al. 2007; Quaiser et al. 2008; Delmas et al. 2009, 2013). In eukaryotes, MRN also processes newly replicated telomeres and DSBs that are blocked by DNA hairpin structures or by proteins, such as Ku and the meiotic recombination factor Spo11 (Lobachev et al. 2002; Connelly et al. 2003; Neale et al. 2005; Bonetti et al. 2010; Mimitou and Symington 2010; Langerak et al. 2011).
How MR(N) functions as a DNA end sensor and processing factor is still poorly understood. Although Ku forms a ring structure with DSB-binding affinity in the nanomolar range (Fig. 1) (Blier et al. 1993; Walker et al. 2001), readily explaining how it acts as a DSB sensor, we have not yet arrived at a model that explains the mechanism of DSB detection by MR(N). Many bulk biochemistry experiments on MRN or MR homologs show a relatively moderate DNA-binding affinity in the high nanomolar to micromolar range and, in general, no clear binding specificity for DNA ends (e.g., Lee et al. 2003; Möckel et al. 2012). However, recent single-molecule fluorescence resonance energy transfer (FRET) analysis of human MRN determined an extraordinarily high DNA-binding affinity in the picomolar range (Cannon et al. 2013). This discrepancy may be caused by differing experimental conditions. MR(N) is intrinsically able to form large macromolecular assemblies in vitro (de Jager et al. 2001), and the ratio of higher-order to lower-order multimers of MR(N) might influence its affinity to DNA. This relationship may partly explain the apparent involvement of the Rad50 coiled-coil domain in high affinity DNA binding, as this domain mediates MR(N) multimerization (Lee et al. 2013).
During the last decade, a substantial number of high- and low-resolution structural studies of MR and MRN components have led to plausible models for MR and MRN complexes from different domains of life (Fig. 1). MR or MRN form large bipolar complexes with globular heads that harbor the nucleotide-binding domains (NBDs) of Rad50 and the nuclease domain of Mre11 (Connelly et al. 1998; Anderson et al. 2001; de Jager et al. 2001; Hopfner et al. 2001). The Mre11 nuclease dimerizes and forms the center of the head module (Hopfner et al. 2001; Williams et al. 2008; Das et al. 2010; Park et al. 2011). Each Mre11 protomer binds one Rad50 coiled-coil domain near the Rad50 NBD, generating a conserved M2R2 architecture (Hopfner et al. 2001; Lammens et al. 2011; Lim et al. 2011; Limbo et al. 2012). Prokaryotic Mre11 binds to Rad50 through a carboxy-terminal helix-loop-helix motif (Fig. 2A) (Lammens et al. 2011; Lim et al. 2011; Möckel et al. 2012). The interaction of eukaryotic Mre11 and Rad50 has not been described on a structural level yet. However, structural information is available for the interaction of S. pombe Mre11 with Nbs1, which binds to the Mre11 nuclease dimer through a conserved motif near the carboxyl terminus of Nbs1 (Schiller et al. 2012).
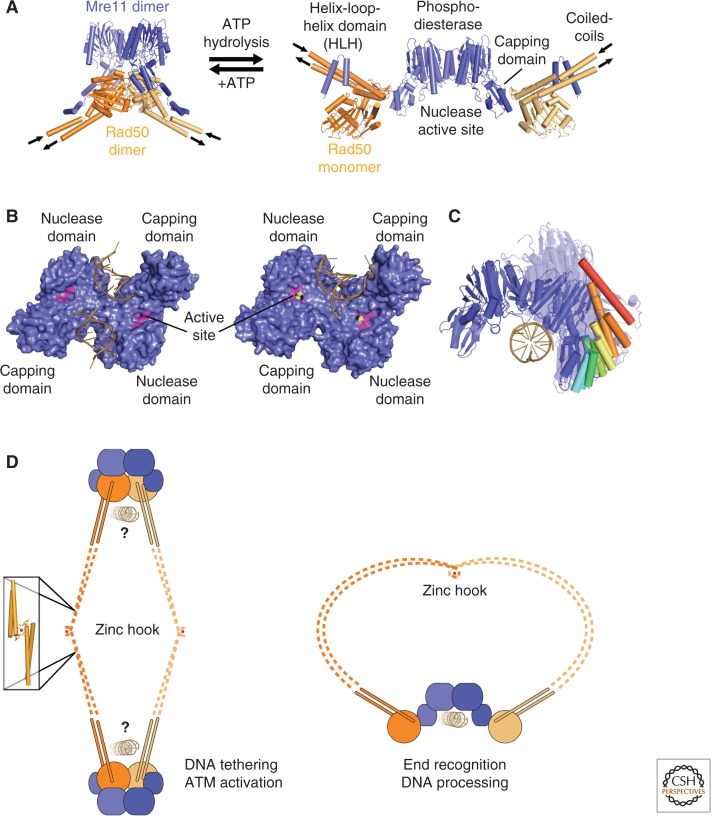
Figure 2.
The Mre11 nuclease and its regulation by Rad50. (A) Structure of the ATP-bound and ATP-free T. maritima MR complex. The PDB codes are 3QG5 and 3THO (Lammens et al. 2011; Möckel et al. 2012). (B) Comparison of Mre11-DNA structures: the surface of the Mre11 dimer (blue) bound to synaptic DNA (left) and branched DNA (right). In the right structure, the active site (magenta) coordinates two manganese ions (yellow). The PDB codes are 3DSC (synaptic DNA) and 3DSD (branched DNA, Williams et al. 2008). (C) Mre11 structure comparison: dimeric crystal structures are aligned onto the left monomer of P. furiosus Mre11 (blue) (PDB code is 1S8E, Arthur et al. 2004). For clarity, the overlaid monomers are not depicted, the right monomers are transparent, and the first α-helix from the capping domain is marked from blue to red to highlight the differences. DNA (sand) indicates the accessible nuclease active site. The PDB codes are 1II7 (Hopfner et al. 2001), 3DSD, 3DSC (Williams et al. 2008), 4HD0 (Limbo et al. 2012), 3AUZ, 3AV0 (Lim et al. 2011), 3THO, 3THN (Möckel et al. 2012), 3QG5 (Lammens et al. 2011), 2Q8U (Das et al. 2010), 4FBQ, 4FBW, 4FBK, and 4FCX (Schiller et al. 2012). (D) MR model for DNA tethering and processing: Mre11 (blue) in complex with Rad50 (orange) forms intercomplex (left) and intracomplex (right) interactions through the zinc hook (zinc ion, red).
The Mre11 Nuclease
Mre11 interacts with both Rad50 and Nbs1 and can be envisioned as the core of the MRN complex. Crystal structures of Mre11 homologs from all three domains of life emphasize the high structural conservation of the amino-terminal Mre11 domain and a universally conserved dimer architecture (Fig. 2B,C) (Hopfner et al. 2001; Arthur et al. 2004; Williams et al. 2008; Das et al. 2010; Lammens et al. 2011; Lim et al. 2011; Limbo et al. 2012; Möckel et al. 2012; Schiller et al. 2012; Liu et al. 2014). The functional importance of Mre11 dimerization is highlighted by findings that mutations of the yeast Mre11 dimer interface phenocopy an mre11 knockout (Williams et al. 2008; Schiller et al. 2012). The conserved amino-terminal domain of Mre11 consists of a phosphoesterase domain and an adjacent capping domain (Fig. 2B). The phosphoesterase active site coordinates two manganese ions, which are essential for exonuclease and ssDNA endonuclease activities (Trujillo et al. 1998; Hopfner et al. 2001).
The Mre11 dimer can directly bind and bridge two DNA ends in vitro (Fig. 2B) (Chen et al. 2001; Williams et al. 2008; Ghodke and Muniyappa 2013), a function that could be important in the context of HR and end-joining reactions (Reis et al. 2012). It is also known that the carboxyl terminus of eukaryotic Mre11 contains additional DNA-binding sites. One site maps to a region adjacent to the capping domain and is crucial for DSB-repair functions. A second DNA-binding motif at the carboxyl terminus of Mre11 was shown to be essential for DSB formation and spore viability in meiosis in S. cerevisiae (Furuse et al. 1998; Usui et al. 1998). Metazoan Mre11 homologs contain, in addition, a glycine/arginine-rich (GAR) motif, which is important for DNA binding and nuclease activity in vitro and localization to DSBs in vivo (Dery et al. 2008).
Comparison of all published structures reveals that the Mre11 dimer angle is not fixed, but it shows a large pivot angle range of one protomer with respect to the other (Fig. 2C). The observed variation of the dimer angle is not necessarily species specific, as S. pombe Mre11, for instance, was crystallized in very different dimer angles in the presence and absence of Nbs1 (Schiller et al. 2012). There might be a correlation between the Mre11 dimer angle and different binding states of Rad50, DNA, and Nbs1. Thus, the observed conformational flexibility might be an important functional aspect that should be addressed in future studies. An exceptional and somewhat surprising case is that of human Mre11, which was crystallized as a dimer cross-linked by an unexpected disulfide bond that leads to an unusual dimer interface and abolishes flexibility (Park et al. 2011).
At present, we have some basic understanding of the interaction of Mre11 with DNA, but important questions remain open. The metal-binding site with its conserved dimetal coordinating histidines readily explains the preference for manganese over magnesium for the 3′ exonuclease. However, P. furiosus Mre11 was also shown to possess magnesium-dependent endonuclease activity that promotes 5′ strand resection, the structural features of which remain elusive so far (Hopkins and Paull 2008). Moreover, our understanding of the molecular mechanism of DNA processing by Mre11 is still limited by the lack of a structure of Mre11 bound to a transition state DNA substrate.
The Rad50 Coiled-Coils
Arguably, the most distinguished yet most poorly understood structural feature of the MRN complex is the long coiled-coil extensions of Rad50. They emerge from the NBDs of Rad50 and carry the universally conserved “zinc-hook” dimerization motif at their apices (Fig. 1) (Hopfner et al. 2002). Two zinc hooks can dimerize by jointly coordinating a zinc ion via four invariant cysteines, two from each zinc hook (Fig. 2D) (Hopfner et al. 2002). In vitro, this dimerization can tether different MRN complexes or help to form supramolecular assemblies to cross-link DNA (de Jager et al. 2001; Hopfner et al. 2002), a feature that may explain the ability of MRN to aggregate DNA in Xenopus cell extracts (Costanzo et al. 2004).
Although the lengths of the coiled-coils are rather conserved between more closely related phylogenetic taxa, they can considerably vary between the different domains of life (Fig. 1). Studies in yeast have shown that the zinc hooks are critical for the function of the complex, but can be partly substituted by dimerization domains of a different type (Wiltzius et al. 2005) or can be compensated for by higher concentrations of MRN in the context of ATM activation (Lee et al. 2013). However, reduction of the length of the coiled-coil dramatically impairs functionality of the MRN complex (Hohl et al. 2011; Deshpande et al. 2014). It is interesting to note that yeast MRN is impaired when the length of the Rad50 coiled-coils is reduced to that of the bacteriophage protein. These results suggest that the dimensions of the Rad50 coiled-coil regions seem to be functionally relevant, but the mechanistic requirements differ strongly between phylogenetic kingdoms and phages. However, care should be taken in the interpretation of these results and the design of such studies, as it is difficult to alter the length of coiled-coil domains without affecting their proper assembly or the orientation of the zinc hooks because of the helical nature of coiled-coils.
Scanning force microscopy (SFM) shows that the coiled-coil domains of Rad50 are organized into segments with flexible hinges that seem to coincide with regions of lower coiled-coil propensity (van Noort et al. 2003; de Jager et al. 2004). Because of this flexibility, two coiled-coil domains can form both inter- and intracomplex interactions, mediated by the dimerization of two zinc-hook motifs (Fig. 2D) (de Jager et al. 2001; Hopfner et al. 2001, 2002; Moreno-Herrero et al. 2005). Importantly, the recent structure of a small, Rad50-like prokaryotic DSB repair factor, RecN, described, for the first time, an atomic model for a full Rad50/SMC/RecN-type structure, assembled from overlapping, crystallographically resolved fragments (Fig. 1) (Pellegrino et al. 2012). This RecN dimer model illustrates the segmental nature of the coiled-coils, but at the same time, it suggests that the coiled-coil domain is overall rather stiff (Fig. 1).
Integrative Model for MR Mechanism
The ATP-binding and hydrolysis motifs of Rad50 are functionally critical elements of MRN. The NBDs of Rad50 dimerize in response to ATP binding, and studies with isolated NBDs show that Rad50 binds DNA in this ATP-engaged conformation (Hopfner et al. 2000b). ATP binding to the NBDs is also important for other functions of the complex such as activation of DNA damage checkpoint regulator ATM (Lee et al. 2013; Deshpande et al. 2014). Recent structural analysis on Mre11-Rad50NBD head complexes revealed that the NBDs of Rad50 are far apart in the absence of ATP, allowing DNA to access the Mre11 nuclease active sites (Fig. 2A) (Lammens et al. 2011). In the presence of ATP, however, the two NBDs dimerize and bind into the DNA-binding/nuclease cleft of the Mre11 dimer (Lim et al. 2011; Möckel et al. 2012; Deshpande et al. 2014). In this conformation, the two active sites of the Mre11 dimer are blocked, at least for binding of double-stranded DNA (dsDNA). These structural studies are consistent with reports that ATP binding to Rad50 negatively regulates the processive 3′ dsDNA exonuclease and dsDNA endonuclease activity (but not the ssDNA endonuclease activity) of Mre11 (Herdendorf et al. 2011; Lim et al. 2011; Majka et al. 2012; Deshpande et al. 2014). The closed, ATP-bound conformation is also the conformation that activates ATM (Lee et al. 2013; Deshpande et al. 2014). Thus, a model may be formulated that was confirmed in a very recent study (Deshpande et al. 2014): The closed MR(N) complex is involved in ATM activation and DSB recognition or tethering, whereas the open complex after ATP hydrolysis is involved in DNA processing (Fig. 2D). It is yet unclear, however, how MRN binds DNA in the closed conformation, in which the Mre11 dsDNA-binding sites are blocked. We also do not know how Rad50 interacts with DNA.
The nature of supramolecular structures of MR and MRN that involve additional interactions mediated by the coiled-coils still needs to be resolved. Several different architectures are conceivable and may play roles in recombination and end joining. Using scanning force microscopic analysis of human MRN, DNA binding was shown to cause a shift from intra-MRN to inter-MRN hook–hook interactions through a mesoscale conformational change (Fig. 2D) (Moreno-Herrero et al. 2005). Therefore, the formation of higher-order structures could be directly coupled to DNA binding. The situation may be different for the rather short coiled-coil structures of the bacteriophage Rad50 orthologs, which leave little room for intramolecular coiled-coil interactions; thus, more work is needed to functionally dissect and validate different superstructures.
Nbs1
The eukaryote-specific subunit of the MRN complex, Nbs1 (or Xrs2 in S. cerevisiae), has multiple functions. It was found to stimulate DNA binding and unwinding of MRN (Paull and Gellert 1999; Trujillo et al. 2003) and is necessary for the nuclear localization of Mre11 and Rad50 (Carney et al. 1998; Desai-Mehta et al. 2001; Tsukamoto et al. 2005). Nbs1 recruits and helps to activate the DNA damage checkpoint regulator ATM/Tel1p (Nakada et al. 2003; Falck et al. 2005; You et al. 2005; Berkovich et al. 2007). Although MR alone seems to be able to interact with ATM in vitro (Costanzo et al. 2004; Lee and Paull 2004; Lee and Paull 2005), the Nbs1 carboxyl terminus was shown to interact with and activate ATM through an acidic patch and a FXF/Y motif (Falck et al. 2005; You et al. 2005). A carboxy-terminal 147-amino-acid fragment of Nbs1 carrying these two motifs was sufficient to restore ATM activation in an Nbs1-depleted Xenopus egg extract (You et al. 2005). In addition, the carboxyl terminus of Nbs1 was found to be necessary for control of cell cycle arrest and apoptosis signals in a mouse model (Stracker et al. 2007).
Nbs1 comprises a folded amino-terminal region and a carboxy-terminal part predicted to be of low structural order (Williams et al. 2009). Crystal structures of the amino-terminal folded region revealed a rigid structure that consists of a fork-head-associated (FHA) domain and tandem BRCA1 carboxy-terminal (BRCT) domains (Lloyd et al. 2009; Williams et al. 2009). FHA and BRCT domains have been shown to recognize phosphoproteins (Durocher and Jackson 2002; Yu et al. 2003). In Nbs1, these domains serve as a recruitment platform for various DSB repair factors such as mediator of DNA damage checkpoint protein 1 (MDC1), Bloom syndrome mutated (BLM), breast cancer 1 (BRCA1), CtBP-interacting protein (CtIP), and phosphorylated histone H2AX (via MDC1) (Fig. 1) (Wang et al. 2000; Burma et al. 2001; Kobayashi et al. 2002; Chapman and Jackson 2008; Chen et al. 2008; Melander et al. 2008; Spycher et al. 2008; Wu et al. 2008). At least in the case of MDC1, both FHA and BRCT domains participate in an interdependent fashion (Lloyd et al. 2009; Hari et al. 2010).
Because of its flexible nature, only limited structural information is available for the carboxy-terminal region of Nbs1. Nbs1 binds to Mre11 through a conserved NFKxFxK motif in this carboxy-terminal region (Desai-Mehta et al. 2001; Tauchi et al. 2001; You et al. 2005; Schiller et al. 2012). Significantly, the crystal structure of S. pombe Mre11 in complex with a carboxy-terminal fragment of Nbs1 showed that this peptide binds across the Mre11 dimer and breaks its symmetry (Schiller et al. 2012). Whether this binding has only the function to tether Mre11 to Nbs1 or—as the peculiar interaction at the Mre11 dimer axis may indicate—is functionally linked to Mre11-Rad50 conformations should be subject of future studies. It also remains to be clarified how this apparently asymmetric binding translates into the stoichiometry of the MRN complex (2:2:2 or 2:2:1).
Mutations in Mre11-Rad50-Nbs1 in Human Disease
Although knockouts of MRE11, RAD50, and NBS1 are lethal in mice (Luo et al. 1999; Zhu et al. 2001; Buis et al. 2008), there are hypomorphic mutations of these genes that are associated with a set of related but phenotypically distinct syndromes such as ataxia-telangiectasia-like disease (ATLD), Nijmegen breakage syndrome (NBS), and NBS-like disorder (NBSLD). These diseases are related to ataxia telangiectasia (A-T), which is caused by mutations in ATM (Savitsky et al. 1995). All three MRN-associated syndromes and A-T share phenotypes on a cellular level, but patients differ with respect to the extent of neurological, immunological, and cancer predisposition disorders. Whereas NBS and NBSLD lead to microcephaly, A-T and ATLD are associated with neurodegeneration (Carney et al. 1998; Varon et al. 1998; Stewart et al. 1999; Maser et al. 2001; Waltes et al. 2009; Matsumoto et al. 2011).
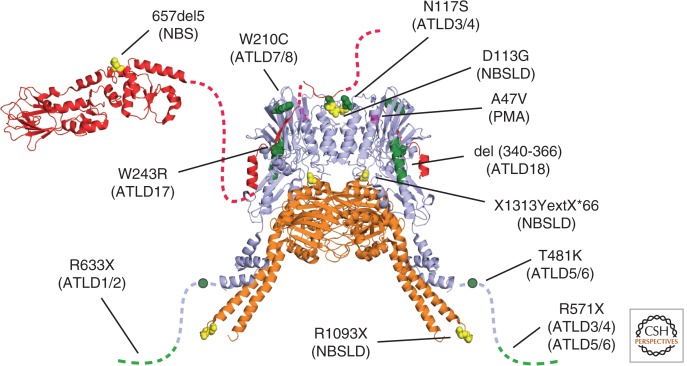
Presently, the literature describes 18 cases of ATLD and one case of NBSLD that were all linked to mutations in the MRE11 gene and one NBSLD patient with two RAD50 mutations (Hernandez et al. 1993; Stewart et al. 1999; Pitts et al. 2001; Delia et al. 2004; Fernet et al. 2005; Uchisaka et al. 2009; Matsumoto et al. 2011; Palmeri et al. 2013). The availability of atomic structures of eukaryotic Mre11 and Nbs1 and prokaryotic Rad50 and the high degree of conservation of MRN allow us to map the underlying mutations onto a structural model of the MRN complex (Fig. 3). Most mutations described so far, apart from truncation mutants, map to the interface between Nbs1 and Mre11. As this interface is quite extended, point mutations reduce, but do not abolish, the interaction between Nbs1 and Mre11, explaining their hypomorphic nature. Functional analysis of some mutations by mutating corresponding conserved residues in S. cerevisiae MRX showed that an ATLD-mimicking mutation did impair mitotic repair functions solely by lowering the nuclear concentration of MRX (Schiller et al. 2012). In addition, telomere maintenance was affected, suggesting a defect in Tel1/ATM activation. For another ATLD-mimicking mutation, a study in S. pombe showed that DSB repair was affected, but not Tel1/ATM activation (Limbo et al. 2012). This situation is somewhat surprising because ATLD is similar to A-T, which is caused by inactivation of ATM. Very recently, progressive myoclonic ataxia (PMA) was also linked to an MRE11 mutation that maps to the surroundings of the Nbs1–Mre11 interface (Miyamoto et al. 2013).
Figure 3.
MRN and human disease. Mapping of MRN mutations found in human disorders onto a model of MRN (model and color code from Fig. 1). ATLD, NBS/-LD, and PMA mutations are indicated in green, yellow, and lilac, respectively.
Further work is thus necessary to correlate the molecular defects in MRN with the observed disease phenotypes. However, the structural studies on the conformational and functional states of MRN will now allow a more detailed structure–function correlation. The mutations may affect these distinct states of MRN and may lead to partial separation of function, which may explain how different disease phenotypes such as NBS and ATLD can result from mutations in a single complex.
RESECTION
Once a DNA DSB has been recognized, 5′-3′ resection of the DNA ends may proceed, which requires a 5′-3′ nuclease and, in most pathways, a helicase. Although this principle holds true for all three domains of life, resection and the initiation thereof are governed by different machineries with conservation limited to single domains. In bacteria, the multisubunit complexes RecBCD and AddAB are stand-alone machineries that recognize DSBs, initiate resection, and perform long-range resection in a highly processive way (for an excellent recent review, see Wigley 2013). Under certain circumstances, an alternative resection pathway may take over that involves the nuclease RecJ and the helicase RecQ (Handa et al. 2009). In archaea, the MR complex identifies DSBs and initiates resection, but a complex comprising the nuclease NurA and the helicase HerA executes long-range resection (Hopkins and Paull 2008; Blackwood et al. 2012). There is evidence, however, that NurA-HerA may form a larger resection complex together with MR (Quaiser et al. 2008). In eukaryotes, the MRN complex initiates resection together with the protein CtIP (Limbo et al. 2007; Mimitou and Symington 2008). Eukaryotic long-range resection has been found to follow partly redundant pathways that involve either the processive nuclease Exo1 or the complex of the nuclease/helicase DNA2 and the RecQ-like helicase Bloom syndrome mutated (BLM, Sgs1 in yeast) (Gravel et al. 2008; Mimitou and Symington 2008; Nimonkar et al. 2008; Zhu et al. 2008). In the following section, we will describe the initiation of resection in eukaryotes, followed by a discussion of the recent advances of our structural understanding of the Exo1, DNA2/BLM, and NurA/HerA pathways.
Initiation of Resection in Eukaryotes
In eukaryotes, initial resection of DSBs requires the MRN complex and CtIP. The precise biochemical function of CtIP is still controversial. CtIP was first characterized as an interaction factor of the transcriptional repressor CtBP, RB1, and the DNA repair and checkpoint protein BRCA1 (Fusco et al. 1998; Schaeper et al. 1998; Wong et al. 1998; Yu et al. 1998). Putative orthologs of CtIP are found in most eukaryotic species, although sequence identity is limited to small regions at the amino and carboxyl termini. CtIP orthologs (Sae2 in S. cerevisiae and Ctp1 in S. pombe) also vary considerably in length and may have diverged in their exact function, for example, with regard to the interaction with other proteins. Nonetheless, both Sae2 and Ctp1 were reported to play roles in initial DNA end resection similar to vertebrate CtIP (McKee and Kleckner 1997; Prinz et al. 1997; Lengsfeld et al. 2007; Limbo et al. 2007; Akamatsu et al. 2008; Nicolette et al. 2010).
Two sequence motifs are conserved between CtIP orthologs from most species. One motif is a predicted coiled-coil region at the amino terminus, which appears to mediate dimerization of CtIP and Sae2, a prerequisite to its functionality (Dubin et al. 2004; Kim et al. 2008; Wang et al. 2012). The second conserved region maps to the carboxyl terminus. It harbors a phosphorylation site (T847 in human and S267 in S. cerevisiae, but absent in S. pombe Ctp1 and some other fungi) that is phosphorylated by CDK to initiate resection (Huertas et al. 2008; Huertas and Jackson 2009). The carboxyl terminus also contains a functionally important CxxC motif (absent in S. cerevisiae Sae2) (Limbo et al. 2007; Akamatsu et al. 2008), mutations of which lead to defects in fission yeast DSB repair almost as severe as a ctp1 knockout. However, the biochemical function of this motif remains to be characterized.
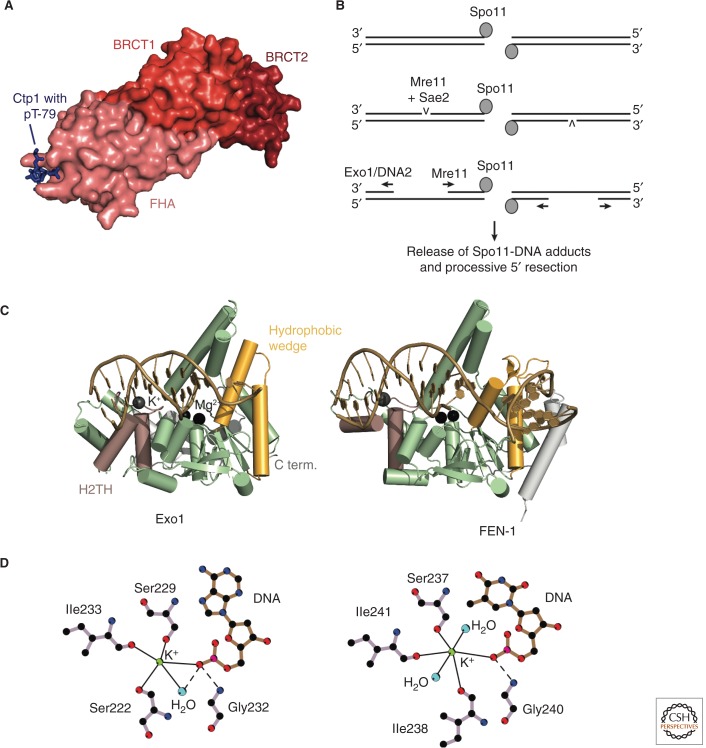
Mammalian CtIP was shown in several studies to physically interact with MRN (Sartori et al. 2007; Chen et al. 2008; Yuan and Chen 2009). In the case of S. pombe Ctp1, two crystal structures illustrate how the amino-terminal FHA domain of Nbs1 binds to a phosphorylated Thr-Asp motif in Ctp1 (Fig. 4A) (Lloyd et al. 2009; Williams et al. 2009). This motif in Ctp1 is phosphorylated in a cell cycle–dependent manner by kinase CK2 (Dodson et al. 2010). Recently, a direct interaction was also reported for recombinant S. cerevisiae MRX and Sae2 (Ghodke and Muniyappa 2013).
Figure 4.
Resection initiation and the Exo1 resection pathway. (A) Structure of an Nbs1-Ctp1 complex from S. pombe (PDB code is 3HUF, Williams et al. 2009). Nbs1 surface with highlighted FHA (salmon), BRCT1 (red), and BRCT2 (dark red) domains. Peptide from Ctp1 (blue) with phosphorylated T79 bound to the FHA domain. (B) Model for bidirectional resection at meiotic DSBs by Mre11 and Exo1/Sgs1-Dna2. A study of meiotic resection in S. cerevisiae suggests an endonucleotic cleavage of the 5′ strand by Mre11/Sae2 at a distance of up to 300 bp away from the Spo11-blocked DNA end. The nicked DNA can then be processed bidirectionally by Mre11 in the 3′-5′ direction and by Exo1 or Sgs1-Dna2 in the 5′-3′ direction (Garcia et al. 2011). (C) Comparison of human Exo1 and FEN-1. Features discussed in the text are highlighted and labeled for Exo1. The PDB codes are 3QEA (Exo1, Orans et al. 2011) and 3Q8K (FEN-1, Tsutakawa et al. 2011). (D) The helix-two-turn-helix (H2TH)-K+ motif in Exo1 (left) and FEN-1 (right). Straight lines indicate metal coordination, and dashed lines indicate hydrogen bonds. Only residues involved in K+-coordination and binding of the K+-coordinating DNA base are shown. The scheme was drawn using LIGPLOT (Laskowski and Swindells 2011). C term., carboxy terminal.
On the basis of studies in budding yeast, Sae2/Ctp1/CtIP has been suggested to initiate resection at DSB ends together with MRN by removing a short stretch of 50 to 100 bp from the 5′ strand. Then, processive nucleases and nuclease–helicase complexes like Exo1 and Sgs1-Dna2 take over to resect the 5′ strand up to the level of several kilobases (Mimitou and Symington 2008; Zhu et al. 2008). It is unclear, however, what defines the number of nucleotides to be removed by MRN and CtIP.
Priming endonucleolytic cleavage by MRN and CtIP may help to process DNA ends blocked by chemical modifications or by proteins and also offers a way to prevent uncontrolled resection and hyperrecombination. Blocked DNA ends occur in meiosis or during abortive topoisomerase reactions (Hartsuiker et al. 2009; Longhese et al. 2009). The MRN(X) complex and Sae2/CtIP may indeed be dispensable for the resection of “clean” DNA ends, as shown for HO-endonuclease and I-SceI-induced DSBs in yeast (Llorente and Symington 2004; Westmoreland and Resnick 2013). In contrast to a model, in which MRN(X) and Sae2/CtIP start resection directly at a DNA end, newer studies provide an alternative model, in which MRN(X) incises DNA away from the DNA end (Fig. 4B). For yeast meiotic DNA breaks that are covalently bound by Spo11 (Keeney et al. 1997), it was shown that the MRX complex and Sae2 incise this blocked DNA up to 300 bases downstream from the DSB and resect the DNA strand in a 3′-5′ direction toward the break (Fig. 4B) (Garcia et al. 2011). This model reconciles the discrepancy between the 3′-5′ exonuclease activity of Mre11 observed in vitro and the 5′-3′ resection observed in vivo. DNA incision away from the DSB was found for HR also in mitotic mammalian cells (Shibata et al. 2014), suggesting that this mode of action is not limited to meiosis. However, some issues remain unclear. What defines the distance between the meiotic or mitotic break and the endonucleolytic incision? The observed distance could be particular to the experiment, but may also reflect the influence of nucleosomes or the tethering function of the Rad50 coiled-coils. Another open question is how MRN distinguishes the two DNA strands during the endonucleolytic cut so that it processes toward and not away from the break.
The biochemical mechanisms by which Sae2/Ctp1/CtIP promote DSB resection are not understood and may differ between species. Although S. cerevisiae Sae2 shows in vitro endonuclease activity on ssDNA and degrades hairpin DNA structures cooperatively with MRN (Lengsfeld et al. 2007), human CtIP seems to lack nuclease activity, but stimulates the ssDNA endonuclease activity of Mre11 (Sartori et al. 2007). Structural data for Sae2/Ctp1/CtIP or its interaction with MRN(X) so far remain elusive. Thus, many questions are still unanswered: (1) Where is the active site in S. cerevisiae Sae2 located? (2) Does it represent a new class of nuclease domains that is absent in vertebrate CtIP? (3) How can this protein stimulate and regulate the nuclease activities of Mre11 in pathways such as mitotic and meiotic HR, telomere maintenance, and MMEJ?
Exonuclease 1 Resection Pathway
The long-range resection nuclease exonuclease 1 (Exo1, in humans, also Hex1) was first described as a 5′-3′ exonuclease from S. pombe (Szankasi and Smith 1992) and then found in S. cerevisiae (Fiorentini et al. 1997; Tishkoff et al. 1997) and humans (Schmutte et al. 1998; Tishkoff et al. 1998; Wilson et al. 1998). Besides its involvement in resection (Tsubouchi and Ogawa 2000; Mimitou and Symington 2008; Zhu et al. 2008; Nicolette et al. 2010), Exo1 has major roles in mismatch repair (Szankasi and Smith 1995; Genschel et al. 2002) and telomere maintenance (Wu et al. 2012). During resection, human Exo1 is stimulated by BLM, MRN, and the replication protein A (RPA), as shown by experiments using purified proteins (Nimonkar et al. 2008, 2011). However, this situation may be different in yeast (Cannavo et al. 2013).
Exo1 belongs to the XPG/Rad2 and FEN-1 family of structure-specific nucleases, a class of metalloenzymes (Shen et al. 1997; Lee and Wilson 1999; Orans et al. 2011). All members of this family share a conserved amino-terminal nuclease domain (amino acids 1–350 in human Exo1), whereas the carboxyl terminus (the remaining 500 amino acids) is divergent. Human Exo1 is dependent on Mg2+, and significantly less active in the presence of Mn2+ (Lee and Wilson 1999). For many years, structural information was limited to crystal structures of the paralog flap endonuclease 1 (FEN-1) (Hosfield et al. 1998; Hwang et al. 1998; Matsui et al. 2002; Chapados et al. 2004; Feng et al. 2004; Sakurai et al. 2005; Doré et al. 2006; Devos et al. 2007), which, among other roles, removes Okazaki fragments during replication (reviewed in Balakrishnan and Bambara 2013). However, these structures were incomplete with regard to the metal center or DNA complexation. Only recently, the crystal structures of human Exo1 and FEN-1, each in complex with a DNA substrate, were reported (Fig. 4C) (Orans et al. 2011; Tsutakawa et al. 2011).
The Exo1 fold comprises a central, twisted β-sheet surrounded by α-helices and is structurally very similar to FEN-1 and related endonucleases (Orans et al. 2011). In the crystal structure, a bound DNA substrate is simulated using a short DNA duplex with a 3′ single-strand extension. The double-stranded part of the DNA interacts with Exo1 only at two points that are set one turn apart. A helix-two-turn-helix (H2TH) motif binding a K+ makes nonspecific bonds with the nonsubstrate strand (Fig. 4C,D). At the active site, two structurally conserved helices form a hydrophobic wedge that drives the nonsubstrate strand into a sharp bend away from the nuclease. The 5′ end of the substrate strand is led to the active site, which consists of two divalent cations that are coordinated by five conserved acidic residues and a conserved lysine and arginine. A remarkable feature is the fraying of the duplex DNA. As a consequence, the substrate strand becomes a single strand that exposes its scissile bond to the metal center. It was proposed that one of the metals activates a water molecule that can attack the scissile bond, whereas the other metal stabilizes the leaving group (Beese and Steitz 1991; Steitz and Steitz 1993; Orans et al. 2011).
Many of these structural features are conserved between the paralogs Exo1 and FEN-1 despite different substrate specificities (Fig. 4C) (Orans et al. 2011; Tsutakawa et al. 2011). Exo1 is primarily an exonuclease at DNA nicks, FEN-1 removes DNA flaps, and other family members such as XPG or GEN cut at DNA bubbles or Holliday junctions, respectively (Tsutakawa and Tainer 2012). Common to all these DNA structures is a nick or gap, and the insights gained from the Exo1 and FEN-1 structures allow the formulation of a single, common mechanism for their processing by FEN-1 family members (for an in-depth discussion of the two crystal structures and their implications, see Grasby et al. 2012; Tsutakawa and Tainer 2012). The presence of a DNA nick or gap is required by the hydrophobic wedge that induces a sharp bend into the template DNA strand and thus prevents the processing of dsDNA. Together with the wedge, the H2TH/K+ motif (Fig. 4D) orientates the substrate strands toward the active site metal center. This potassium ion is absent in FEN-1 crystal structures that lack DNA (e.g., Chapados et al. 2004). A similar configuration was observed in the DNA polymerase β, where a K+ is involved in binding of a helix-hairpin-helix motif to DNA and was suggested to support processivity (Pelletier et al. 1996). Access to the active site is restricted to ssDNA, again excluding a continuous dsDNA. The ssDNA is generated by melting of two residues of the substrate strand.
Still under discussion is the interaction between FEN-1 paralogs and the substrate ssDNA upstream of the incision. A threading mechanism has been postulated that requires the threading of the ssDNA through a helical arch that is disordered in the absence of bound DNA (Ceska et al. 1996; Tsutakawa et al. 2011; Tsutakawa and Tainer 2012; Balakrishnan and Bambara 2013). Threading has to be ruled out for a DNA bubble in the case of XPG. Some argue that XPG probably does not have a helical arch (Tsutakawa et al. 2011; Tsutakawa and Tainer 2012), whereas others posit that the ssDNA may circumvent the helical arch and bind on surface grooves, which may extend to all FEN-1-related nucleases, including XPG and GEN (Orans et al. 2011).
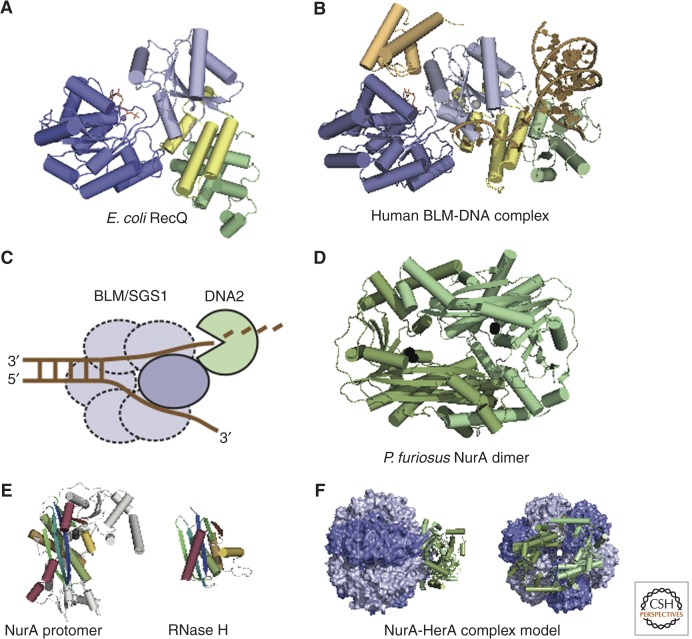
The Sgs1/BLM-DNA2 Resection Pathway
The second main pathway for processive 5′-resection in HR, beside Exo1-mediated resection, depends on the cooperative action of the nuclease DNA2 and the helicase activity of Sgs1 in S. cerevisiae or its functional homolog BLM in vertebrates (Gravel et al. 2008; Mimitou and Symington 2008; Nimonkar et al. 2008; Zhu et al. 2008). Sgs1 or BLM unwind duplex DNA by their 3′-5′ helicase activity. The ssDNA-binding protein, replication protein-A (RPA), then coats ssDNA unwound by Sgs1 and promotes 5′-3′ degradation by Dna2 while inhibiting 3′ to 5′ degradation (Cejka et al. 2010a; Niu et al. 2010; Nimonkar et al. 2011). Recombinant Dna2 and Sgs1 physically interact even in the absence of a DNA substrate, and a similar interaction was also reported for human DNA2 and BLM (Cejka et al. 2010a; Nimonkar et al. 2011). In addition, Sgs1 is part of the Sgs1-Top3-Rmi1 (STR) complex, together with the topoisomerase class I enzyme Topoisomerase III and the regulatory protein Rmi1 (Gangloff et al. 1994; Chang et al. 2005; Mullen et al. 2005). This complex is responsible for dissolution of double Holliday junctions in the late stage of homologous recombination (Cejka et al. 2010b). Top3 and Rmi1 are also important for the resection function of Sgs1. Deletion mutants of all three proteins share similar resection defects in vivo (Zhu et al. 2008), and Top3-Rmi1 also stimulates the 5′-resection capacity of Sgs1-Dna2 in vitro (Cejka et al. 2010a; Niu et al. 2010). A very recent crystal structure of the conserved core of the human TopIIIα-RMI1 complex illustrates how RMI1 might regulate TopIIIα through a long insertion loop that invades the central gate of the toroidal topoisomerase (Bocquet et al. 2014).
Sgs1 and BLM both belong to the RecQ family of helicases. Most prokaryotes and yeasts possess only one or two RecQ homologs (like Sgs1 in S. cerevisiae), whereas in vertebrates multiple homologs are found. For Homo sapiens, five RecQ-like helicases have been described: BLM, WRN, RECQ1, RECQ4, and RECQ5β. All of these helicases play important roles in different pathways of genome maintenance (Chu and Hickson 2009).
The RecQ-like helicases belong to the SF-2 helicase family and share a conserved core, which consists of two RecA-like domains and a carboxy-terminally adjacent RQC (RecQ carboxy-terminal) region (lacking in RecQ4) (Vindigni et al. 2010; Manthei and Keck 2013). The RQC region is needed for strand separation of DNA substrates (Hu et al. 2005; Pike et al. 2009; Kitano et al. 2010). Many RecQ-like helicases, including bacterial RecQ, yeast Sgs1, and vertebrate BLM also possess a helicase and RNaseD carboxy-terminal (HDRC) domain that is important for DNA substrate recognition and translocation (Liu et al. 1999; Bernstein and Keck 2005; Kocsis et al. 2014).
Several atomic resolution structures are now available and yield insights into the functional architecture of these helicases. The crystal structure of Escherichia coli RecQ revealed the principal architecture of the catalytic core (Fig. 5A) (Bernstein et al. 2003). The RecA-like domains and the RQC region, consisting of a zinc-binding motif and a winged helix domain, compose a compact modular arrangement, which is also found in the structure of human RecQ1 (Pike et al. 2009). The HRDC domains from E. coli RecQ, SGS1, and BLM possess a very similar fold. However, they exhibit different DNA substrate specificities. This is reflected in their differing composition of DNA-interacting residues and distinct surface charge distributions (Liu et al. 1999; Bernstein and Keck 2005; Kim and Choi 2010; Sato et al. 2010).
Figure 5.
Structures of nuclease–helicase complexes involved in resection. (A) Structure of the helicase catalytic core of E. coli RecQ bound to ATPγS. The structure consists of the RecA-like helicase domains (dark and light blue) bound to ATPγS (orange) and the RecQ carboxy-terminal region, consisting of the zinc-binding domain (yellow) and a winged-helix domain (green). The PDB code is 10YY (Bernstein et al. 2003). (B) Structure of human BLM helicase in complex with DNA. The color coding is similar to that in A. The HRDC domain and ADP are drawn in orange, and the DNA in brown. The PDB code is 4CGZ. (C) The RecQ-like helicases BLM in vertebrates or Sgs1 in yeast are both cooperating with DNA2 in DSB resection. Sgs1 or BLM unwind dsDNA by their 3′-5′ helicase activity. The ssDNA-binding protein RPA then coats ssDNA unwound by Sgs1 and promotes 5′-3′ degradation by Dna2 (Cejka et al. 2010a; Niu et al. 2010; Nimonkar et al. 2011). The potential of BLM and Sgs1 to form multimers is indicated using dashed lines. (D) Structure of the P. furiosus NurA dimer. The PDB code is 3TAL (Chae et al. 2012). (E) RNase H fold of NurA. A Comparison of a P. furiosus NurA protomer with E. coli RNase H (PDB code 1RNH, Yang et al. 1990). Homologous elements are highlighted using the same color. (F) Model of the NurA-HerA complex. The crystal structure of Sulfolobus solfataricus NurA (PDB code 2YGK, Blackwood et al. 2012) is fitted to a HerA homolog, the conjugation protein TrwB (PDB code 1E9R, Gomis-Ruth et al. 2001).
Very recently, structures of human BLM in complex with partially unwound DNA were determined (PDB code 4CGZ, Fig. 5B) (Swan et al. 2014; O Gileadi, pers. comm.). This structure reveals extensive interactions of the winged helix domain with the upstream dsDNA “substrate” and shows that the β-hairpin wing acts as a DNA-splitting element. The zinc-binding insertion domain functions as single-stranded DNA ratchet, whereas the RecA-like domains binds to the ssDNA “product.” The HRDC domain does not make any DNA contacts but is positioned on top of the nucleotide-binding cleft of the RecA-like domains. It will be interesting to see whether the observed position of the HRDC domain is important for the regulation of BLM helicase activity. Bacterial RecQ and BLM structures share high fold conservation, although the winged-helix domain is positioned differently in both structures, indicating flexibility for this element (Fig 5A,B).
Many RecQ-like helicases including BLM are known to form oligomers, at least in vitro (Fig. 5C) (Karow et al. 1999; Xue et al. 2002; Perry et al. 2006; Vindigni and Hickson 2009). However, a more recent study describes that BLM oligomers dissociate into monomers upon ATP hydrolysis and that only monomeric but not oligomeric BLM displays DNA unwinding activity (Xu et al. 2012a). Thus, it remains an open question which functional role oligomerization of Sgs1 or BLM plays in the context of 5′-strand resection in HR.
Homologs of the nuclease-helicase protein DNA2 are found both in archaea and eukarya but are absent in bacteria, although the bacterial AddAB system bears some structural similarity. Although archaeal Dna2 is only poorly characterized (Higashibata et al. 2003), the eukaryotic protein was found to play crucial roles in several genome maintenance processes beside homologous recombination, including Okazaki fragment processing (Kang et al. 2010) and telomere stabilization (Lin et al. 2013). Initial genetic studies in S. cerevisiae revealed that the nuclease activity of Dna2 is essential in vivo, whereas a helicase-dead mutant strain is viable at lower growth temperatures (Budd et al. 2000). A later study then clarified that the nuclease of Dna2 is responsible for processive 5′ strand resection in DSB repair by HR where it is the second important processive nuclease beside Exo1 (Zhu et al. 2008). Both Exo1 and Dna2 function independently of each other and seem to play redundant roles (Zhu et al. 2008; Cannavo et al. 2013). The nuclease module of Dna2 belongs to the RecB family and maps to the amino terminus of the protein. Remarkably, it was shown to contain an iron–sulfur cluster, which is crucial for both nuclease and ATPase activity (Yeeles et al. 2009; Pokharel and Campbell 2012). This situation is reminiscent of the bacterial DSB resection protein AddB, which possesses a nuclease domain with a 4Fe–4S cluster (Yeeles et al. 2009).
In contrast to its nuclease activity, the helicase activity of Dna2 is dispensable for 5′-strand resection, which was a rather surprising finding. Instead, the unwinding activity of RecQ-like helicases such as Sgs1 or BLM provides the 5′-ssDNA for resection by Dna2 (Cejka et al. 2010a; Niu et al. 2010). The Dna2 helicase exhibits only a weak 5′-3′ unwinding activity on dsDNA and depends on the binding to free DNA ends before acting as a helicase (Bae et al. 2002). However, recently, it was shown that Dna2 is a vigorous 5′-3′ helicase in an S. cerevisiae nuclease dead mutant (Levikova et al. 2013). Apparently, Dna2 depends on the binding to 5′-ssDNA flaps for processive helicase activity, and these flaps are degraded by the Dna2 nuclease domain of the wild-type protein (Levikova et al. 2013). There may be a structural switch in Dna2 that regulates the balance between its nuclease and helicase activities. Atomic resolution structures of Dna2, Sgs1, and their DNA substrates, which are still lacking at the moment, could provide important information to better understand how these key enzymes cooperate to specifically resect 5′-DNA at DSBs.
Resection in Archaea
Archaea, like eukaryotes, use homologs of Mre11-Rad50 for resection. However, for long-range resection, Mre11-Rad50 are joined by two proteins unique to archaea, the helicase HerA and the nuclease NurA (Manzan et al. 2004; Hopkins and Paull 2008; Blackwood et al. 2012; Chae et al. 2012). Archaeal resection is thus similar to the eukaryotic process with regard to common players such as Mre11-Rad50 and the overall principle. In contrast, the archaeal resection machinery is completely distinct from bacterial proteins such as RecBCD, AddAB, or AdnAB, although some archaeal species may have taken up AddAB-like proteins by horizontal gene transfer (Cromie 2009).
The genes nurA and herA are encoded in one operon together with mre11 and rad50 in almost all archaea (Constantinesco et al. 2002,2004; Manzan et al. 2004). NurA is a dimer and has been described as both a 5′ and 3′ exonuclease for dsDNA and ssDNA and an endonuclease for ssDNA (Constantinesco et al. 2002; Hopkins and Paull 2008; Wei et al. 2008,2011; Blackwood et al. 2012; Chae et al. 2012). HerA is a hexameric, ATP-dependent DNA helicase that is activated by and unwinds dsDNA in both the 5′ and 3′ direction (Constantinesco et al. 2004; Manzan et al. 2004; Zhang et al. 2008).
For full activity, NurA and HerA have to form a complex, which is stable in vitro, at least for species such as P. furiosus and S. solfataricus (Hopkins and Paull 2008; Blackwood et al. 2012; Chae et al. 2012). However, this physical interaction may be less stable or absent in Sulfolobus acidocaldarius (Quaiser et al. 2008). The stoichiometry of the complex (if formed) seems to be 2:6, that is, a NurA dimer and a HerA hexamer (Hopkins and Paull 2008; Blackwood et al. 2012; Chae et al. 2012).
The HerA monomers assemble into a hexameric ring, as visualized by electron microscopy analysis (Manzan et al. 2004). Thus, HerA is a typical member of the FtsK class of P-loop ATPases and is likely to have a similar fold (Constantinesco et al. 2004; Iyer et al. 2004). The HerA amino terminus is predicted to comprise a distinct domain that folds into a β-barrel and was called the HAS (HerA-ATP synthase) domain (Iyer et al. 2004).
The crystal structures of two NurA orthologs from P. furiosus and S. solfataricus were recently reported (Fig. 5D) (Blackwood et al. 2012; Chae et al. 2012). The conserved, active domain is of the RNaseH-like fold (Fig. 5E) with nonconserved extensions that make extensive dimer interactions. The overall shape of the NurA dimer is ring-like, with the active sites of each monomer facing each other within the ring pore (Fig. 5D). Conserved acidic residues bind one or two Mn2+ cations in the active site of P. furiosus NurA, depending on the crystallization conditions (Chae et al. 2012). Mutation of these manganese-binding residues completely inactivates NurA (Hopkins and Paull 2008; Wei et al. 2011; Blackwood et al. 2012; Chae et al. 2012). Also, Mg2+ is known to be essential for NurA activity (Constantinesco et al. 2002; Hopkins and Paull 2008; Chae et al. 2012). Thus, the catalytic mechanism of NurA could be similar to that postulated for the RNaseH-like nuclease Argonaute (Wang et al. 2009) or RNase H itself (Nowotny et al. 2005). Structures of these proteins bound to divalent cations and DNA have led to a model in which one cation activates a water for nucleophilic attack on the DNA backbone and the second cation stabilizes the leaving group (Beese and Steitz 1991; Steitz and Steitz 1993; Nowotny et al. 2005; Wang et al. 2009).
The structures of NurA also yielded insight into the cooperative DNA processing of NurA and HerA. The cavity of the NurA ring is positively charged, as expected for a DNA-processing enzyme. However, it holds space only for one or two ssDNA strands, but not for B-form dsDNA (Blackwood et al. 2012; Chae et al. 2012). The interaction interface of NurA and HerA could be mapped to residues on the flat surface of the NurA ring close to the active site, which are bound by the HerA HAS domain (Fig. 5F) (Blackwood et al. 2012). These data imply that the helicase HerA unwinds dsDNA and passes one or both single strands directly on to the NurA dimer.
The NurA structure does not answer the question of whether, in vivo, the NurA/HerA complex digests one or both strands of dsDNA. However, it has been observed that the rate of ATP hydrolysis by NurA/HerA varies with the nature of the DNA substrate. This rate is higher for dsDNA with blunt ends or short overhangs and lower in the presence of longer overhangs (Blackwood et al. 2012). In consequence, the nature of the DNA end may trigger complete digestion of the DNA double strand or the 5′-3′ resection necessary to produce a DNA tail. Blackwood et al. (2012) suggest that the former could be an archaeal defense mechanism against foreign DNA, whereas the latter might rely on the preparation of the DNA by the MR complex. Experimental evidence indeed supports a model in which the MR complex and the NurA/HerA complex cooperate to produce a 3′ overhang that is then bound by RadA (archaeal RecA) for homologous recombination (Hopkins and Paull 2008).
OPEN QUESTIONS AND CONCLUDING REMARKS
The past decade has brought a plethora of new insights into the composition, biochemistry, and regulation of the DSB detection and resection machineries. We now have an inventory of enzymatic activities at DSBs in all three domains of life. Nonetheless, from a mechanistic and also an evolutionary point of view, we are far from understanding the molecular choreography of DSB detection, repair, and resection. Although the bacterial resection machineries, RecBCD and AddAB, are well characterized, the structural nature of the resection machineries in eukaryotes and archaea requires further attention. New developments in electron microscopy and hybrid methods in structural biology may help to better understand the interaction architectures of these complexes and the interplay of different nuclease, helicase, and topoisomerase activities. Likewise, despite progress over the last years, the mechanism of the MR(N) complex in DNA end processing is still unclear. Several fundamental issues remain unsolved, in particular, the mechanism of DSB detection by MRN, the nature of its cryptic endonuclease activity, and the role and mechanism of the cofactor CtIP/Ctp1/Sae2. Other issues such as identifying the site of the initial endonucleolytic cleavage of DNA ends will require the reconstitution of more complete biochemical systems. It will also be important to mechanistically address the resection in chromatin templates and integrate the activities of chromatin modifying enzymes with resection enzymes. Recent studies have begun to look at exactly that and showed in vitro that Sgs1-Dna2 resection requires some nucleosome-free DNA but can then proceed through nucleosomes. In contrast, nucleosomes provide an obstacle for Exo1-based resection that may be lifted by chromatin-remodeling activities (Adkins et al. 2013).
ACKNOWLEDGMENTS
We apologize to all authors whose work we could not cite owing to space limitations. Work in the K.-P.H. laboratory on DNA double-strand breaks is supported by the European Research Council (ERC) advanced grant “ATMMACHINE,” the German Excellence Initiative Research Cluster CIPSM, and German Research Council Program projects SFB646 and GRK1721.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Adkins NL, Niu H, Sung P, Peterson CL 2013. Nucleosome dynamics regulates DNA processing. Nat Struct Mol Biol 20: 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Gomez-Gonzalez B 2008. Genome instability: A mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Akamatsu Y, Murayama Y, Yamada T, Nakazaki T, Tsutsui Y, Ohta K, Iwasaki H 2008. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol Cell Biol 28: 3639–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almond JR, Stohr BA, Panigrahi AK, Albrecht DW, Nelson SW, Kreuzer KN 2013. Coordination and processing of DNA ends during double-strand break repair: The role of the bacteriophage T4 Mre11/Rad50 (MR) complex. Genetics 195: 739–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunugama R, Fishel R 2012. Homologous recombination in eukaryotes. Prog Mol Biol Transl Sci 110: 155–206 [DOI] [PubMed] [Google Scholar]
- Anderson DE, Trujillo KM, Sung P, Erickson HP 2001. Structure of the Rad50-Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J Biol Chem 276: 37027–37033 [DOI] [PubMed] [Google Scholar]
- Arthur LM, Gustausson K, Hopfner KP, Carson CT, Stracker TH, Karcher A, Felton D, Weitzman MD, Tainer J, Carney JP 2004. Structural and functional analysis of Mre11-3. Nucleic Acids Res 32: 1886–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SH, Kim DW, Kim J, Kim JH, Kim DH, Kim HD, Kang HY, Seo YS 2002. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J Biol Chem 277: 26632–26641 [DOI] [PubMed] [Google Scholar]
- Balakrishnan L, Bambara RA 2013. Flap endonuclease 1: A central component of DNA metabolism. Annu Rev Biochem 82: 119–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese LS, Steitz TA 1991. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase: A two metal ion mechanism. EMBO J 10: 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ Jr, Kastan MB 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690 [DOI] [PubMed] [Google Scholar]
- Bernstein DA, Keck JL 2005. Conferring substrate specificity to DNA helicases: Role of the RecQ HRDC domain. Structure 13: 1173–1182 [DOI] [PubMed] [Google Scholar]
- Bernstein DA, Zittel MC, Keck JL 2003. High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J 22: 4910–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood JK, Rzechorzek NJ, Abrams AS, Maman JD, Pellegrini L, Robinson NP 2012. Structural and functional insights into DNA-end processing by the archaeal HerA helicase–NurA nuclease complex. Nucleic Acids Res 40: 3183–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blier PR, Griffith AJ, Craft J, Hardin JA 1993. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem 268: 7594–7601 [PubMed] [Google Scholar]
- Bocquet N, Bizard AH, Abdulrahman W, Larsen NB, Faty M, Cavadini S, Bunker RD, Kowalczykowski SC, Cejka P, Hickson ID, et al. 2014. Structural and mechanistic insight into Holliday-junction dissolution by Topoisomerase IIIα and RMI1. Nat Struct Mol Biol 21: 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Clerici M, Manfrini N, Lucchini G, Longhese MP 2010. The MRX complex plays multiple functions in resection of Yku- and Rif2-protected DNA ends. PLoS ONE 5: e14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Choe W, Campbell JL 2000. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J Biol Chem 275: 16518–16529 [DOI] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276: 42462–42467 [DOI] [PubMed] [Google Scholar]
- Cadet J, Ravanat JL, TavernaPorro M, Menoni H, Angelov D 2012. Oxidatively generated complex DNA damage: Tandem and clustered lesions. Cancer Lett 327: 5–15 [DOI] [PubMed] [Google Scholar]
- Cannavo E, Cejka P, Kowalczykowski SC 2013. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc Natl Acad Sci 110: E1661–E1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Kuhnlein J, Yang SH, Cheng A, Schindler D, Stark JM, Russell R, Paull TT 2013. Visualization of local DNA unwinding by Mre11/Rad50/Nbs1 using single-molecule FRET. Proc Natl Acad Sci 110: 18868–18873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR III, Hays L, Morgan WF, Petrini JH 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell 93: 477–486 [DOI] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC 2010a. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC 2010b. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol 17: 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceska TA, Sayers JR, Stier G, Suck D 1996. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature 382: 90–93 [DOI] [PubMed] [Google Scholar]
- Chae J, Kim YC, Cho Y 2012. Crystal structure of the NurA-dAMP-Mn2+ complex. Nucleic Acids Res 40: 2258–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW 2005. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J 24: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA 2004. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 116: 39–50 [DOI] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP 2008. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep 9: 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X 2008. Cell cycle-dependent complex formation of BRCA1-CtIP-MRN is important for DNA double-strand break repair. J Biol Chem 283: 7713–7720 [DOI] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE 2013. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol 5: a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID 2009. RecQ helicases: Multifunctional genome caretakers. Nat Rev Cancer 9: 644–654 [DOI] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Okely EA, Leach DR 1997. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J Biol Chem 272: 19819–19826 [DOI] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DR 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci 95: 7969–7974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DR 1999. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res 27: 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DR 2003. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amst) 2: 795–807 [DOI] [PubMed] [Google Scholar]
- Constantinesco F, Forterre P, Elie C 2002. NurA, a novel 5′-3′ nuclease gene linked to rad50 and mre11 homologs of thermophilic Archaea. EMBO Rep 3: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinesco F, Forterre P, Koonin EV, Aravind L, Elie C 2004. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res 32: 1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell 8: 137–147 [DOI] [PubMed] [Google Scholar]
- Costanzo V, Paull T, Gottesman M, Gautier J 2004. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol 2: E110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA 2009. Phylogenetic ubiquity and shuffling of the bacterial RecBCD and AddAB recombination complexes. J Bacteriol 191: 5076–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon E, Eykelenboom JK, Lincker F, Jones LH, White M, Okely E, Blackwood JK, Leach DR 2010. E coli SbcCD and RecA control chromosomal rearrangement induced by an interrupted palindrome. Mol Cell 39: 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Moiani D, Axelrod HL, Miller MD, McMullan D, Jin KK, Abdubek P, Astakhova T, Burra P, Carlton D, et al. 2010. Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair. J Mol Biol 397: 647–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell 8: 1129–1135 [DOI] [PubMed] [Google Scholar]
- de Jager M, Trujillo KM, Sung P, Hopfner KP, Carney JP, Tainer JA, Connelly JC, Leach DR, Kanaar R, Wyman C 2004. Differential arrangements of conserved building blocks among homologs of the Rad50/Mre11 DNA repair protein complex. J Mol Biol 339: 937–949 [DOI] [PubMed] [Google Scholar]
- Delia D, Piane M, Buscemi G, Savio C, Palmeri S, Lulli P, Carlessi L, Fontanella E, Chessa L 2004. MRE11 mutations and impaired ATM-dependent responses in an Italian family with ataxia-telangiectasia-like disorder. Hum Mol Genet 13: 2155–2163 [DOI] [PubMed] [Google Scholar]
- Delmas S, Shunburne L, Ngo HP, Allers T 2009. Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PLoS Genet 5: e1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas S, Duggin IG, Allers T 2013. DNA damage induces nucleoid compaction via the Mre11-Rad50 complex in the archaeon Haloferax volcanii. Mol Microbiol 87: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dery U, Coulombe Y, Rodrigue A, Stasiak A, Richard S, Masson JY 2008. A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol Cell Biol 28: 3058–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai-Mehta A, Cerosaletti KM, Concannon P 2001. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol 21: 2184–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Williams GJ, Limbo O, Williams RS, Kuhnlein J, Lee JH, Classen S, Guenther G, Russell P, Tainer JA, et al. 2014. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J 33: 482–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos JM, Tomanicek SJ, Jones CE, Nossal NG, Mueser TC 2007. Crystal structure of bacteriophage T4 5′ nuclease in complex with a branched DNA reveals how flap endonuclease-1 family nucleases bind their substrates. J Biol Chem 282: 31713–31724 [DOI] [PubMed] [Google Scholar]
- Dodson GE, Limbo O, Nieto D, Russell P 2010. Phosphorylation-regulated binding of Ctp1 to Nbs1 is critical for repair of DNA double-strand breaks. Cell Cycle 9: 1516–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doré AS, Kilkenny ML, Jones SA, Oliver AW, Roe SM, Bell SD, Pearl LH 2006. Structure of an archaeal PCNA1-PCNA2-FEN1 complex: Elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res 34: 4515–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin MJ, Stokes PH, Sum EY, Williams RS, Valova VA, Robinson PJ, Lindeman GJ, Glover JN, Visvader JE, Matthews JM 2004. Dimerization of CtIP, a BRCA1- and CtBP-interacting protein, is mediated by an N-terminal coiled-coil motif. J Biol Chem 279: 26932–26938 [DOI] [PubMed] [Google Scholar]
- Durocher D, Jackson SP 2002. The FHA domain. FEBS Lett 513: 58–66 [DOI] [PubMed] [Google Scholar]
- Eykelenboom JK, Blackwood JK, Okely E, Leach DR 2008. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol Cell 29: 644–651 [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 [DOI] [PubMed] [Google Scholar]
- Feng M, Patel D, Dervan JJ, Ceska T, Suck D, Haq I, Sayers JR 2004. Roles of divalent metal ions in flap endonuclease-substrate interactions. Nat Struct Mol Biol 11: 450–456 [DOI] [PubMed] [Google Scholar]
- Fernet M, Gribaa M, Salih MA, Seidahmed MZ, Hall J, Koenig M 2005. Identification and functional consequences of a novel MRE11 mutation affecting 10 Saudi Arabian patients with the ataxia telangiectasia-like disorder. Hum Mol Genet 14: 307–318 [DOI] [PubMed] [Google Scholar]
- Ferretti LP, Lafranchi L, Sartori AA 2013. Controlling DNA-end resection: A new task for CDKs. Front Genet 4: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS 1997. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol 17: 2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frols S, Gordon PM, Panlilio MA, Duggin IG, Bell SD, Sensen CW, Schleper C 2007. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J Bacteriol 189: 8708–8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J 17: 6412–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco C, Reymond A, Zervos AS 1998. Molecular cloning and characterization of a novel retinoblastoma-binding protein. Genomics 51: 351–358 [DOI] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol 14: 8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapud EJ, Sleckman BP 2011. Unique and redundant functions of ATM and DNA-PKcs during V(D)J recombination. Cell Cycle 10: 1928–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SEL, Gray S, Neale MJ 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschel J, Bazemore LR, Modrich P 2002. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem 277: 13302–13311 [DOI] [PubMed] [Google Scholar]
- Ghodke I, Muniyappa K 2013. Processing of DNA double-stranded breaks and intermediates of recombination and repair by Saccharomyces cerevisiae Mre11 and its stimulation by Rad50, Xrs2, and Sae2 proteins. J Biol Chem 288: 11273–11286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409: 637–641 [DOI] [PubMed] [Google Scholar]
- Grasby JA, Finger LD, Tsutakawa SE, Atack JM, Tainer JA 2012. Unpairing and gating: Sequence-independent substrate recognition by FEN superfamily nucleases. Trends Biochem Sci 37: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC 2009. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev 23: 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari FJ, Spycher C, Jungmichel S, Pavic L, Stucki M 2010. A divalent FHA/BRCT-binding mechanism couples the MRE11-RAD50-NBS1 complex to damaged chromatin. EMBO Rep 11: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E, Neale MJ, Carr AM 2009. Distinct requirements for the Rad32Mre11 nuclease and Ctp1CtIP in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell 33: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdendorf TJ, Albrecht DW, Benkovic SJ, Nelson SW 2011. Biochemical characterization of bacteriophage T4 Mre11-Rad50 complex. J Biol Chem 286: 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D, McConville CM, Stacey M, Woods CG, Brown MM, Shutt P, Rysiecki G, Taylor AM 1993. A family showing no evidence of linkage between the ataxia telangiectasia gene and chromosome 11q22-23. J Med Genet 30: 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashibata H, Kikuchi H, Kawarabayasi Y, Matsui I 2003. Helicase and nuclease activities of hyperthermophile Pyrococcus horikoshii Dna2 inhibited by substrates with RNA segments at 5′-end. J Biol Chem 278: 15983–15990 [DOI] [PubMed] [Google Scholar]
- Hohl M, Kwon Y, Galvan SM, Xue X, Tous C, Aguilera A, Sung P, Petrini JH 2011. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat Struct Mol Biol 18: 1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP 2000a. Mre11 and Rad50 from Pyrococcus furiosus: Cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol 182: 6036–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA 2000b. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101: 789–800 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105: 473–485 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418: 562–566 [DOI] [PubMed] [Google Scholar]
- Hopkins BB, Paull TT 2008. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 135: 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen B, Tainer JA 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: Coupling DNA and PCNA binding to FEN-1 activity. Cell 95: 135–146 [DOI] [PubMed] [Google Scholar]
- Hu JS, Feng H, Zeng W, Lin GX, Xi XG 2005. Solution structure of a multifunctional DNA- and protein-binding motif of human Werner syndrome protein. Proc Natl Acad Sci 102: 18379–18384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang KY, Baek K, Kim HY, Cho Y 1998. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat Struct Biol 5: 707–713 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Makarova KS, Koonin EV, Aravind L 2004. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res 32: 5260–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5: a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YH, Lee CH, Seo YS 2010. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit Rev Biochem Mol Biol 45: 71–96 [DOI] [PubMed] [Google Scholar]
- Karow JK, Newman RH, Freemont PS, Hickson ID 1999. Oligomeric ring structure of the Bloom’s syndrome helicase. Curr Biol 9: 597–600 [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kim YM, Choi BS 2010. Structure and function of the regulatory HRDC domain from human Bloom syndrome protein. Nucleic Acids Res 38: 7764–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Vijayakumar S, Reger M, Harrison JC, Haber JE, Weil C, Petrini JH 2008. Functional interactions between Sae2 and the Mre11 complex. Genetics 178: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano K, Kim SY, Hakoshima T 2010. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure 18: 177–187 [DOI] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K 2002. NBS1 localizes to γ-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol 12: 1846–1851 [DOI] [PubMed] [Google Scholar]
- Kocsis ZS, Sarlos K, Harami GM, Martina M, Kovacs M 2014. A nucleotide- and HRDC-domain-dependent structural transition in DNA-bound RecQ helicase. J Biol Chem 289: 5938–5949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer KN, Brister JR 2010. Initiation of bacteriophage T4 DNA replication and replication fork dynamics: A review in the Virology Journal series on bacteriophage T4 and its relatives. Virol J 7: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lam I, Keeney S 2014. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens K, Bemeleit DJ, Möckel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermüller C, Söding J, et al. 2011. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145: 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet 7: e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Swindells MB 2011. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model 51: 2778–2786 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304: 93–96 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554 [DOI] [PubMed] [Google Scholar]
- Lee BI, Wilson DM III 1999. The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem 274: 37763–37769 [DOI] [PubMed] [Google Scholar]
- Lee JH, Ghirlando R, Bhaskara V, Hoffmeyer MR, Gu J, Paull TT 2003. Regulation of Mre11/Rad50 by Nbs1: Effects on nucleotide-dependent DNA binding and association with ataxia-telangiectasia-like disorder mutant complexes. J Biol Chem 278: 45171–45181 [DOI] [PubMed] [Google Scholar]
- Lee JH, Mand MR, Deshpande RA, Kinoshita E, Yang SH, Wyman C, Paull TT 2013. Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. J Biol Chem 288: 12840–12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levikova M, Klaue D, Seidel R, Cejka P 2013. Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc Natl Acad Sci 110: E1992–E2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HS, Kim JS, Park YB, Gwon GH, Cho Y 2011. Crystal structure of the Mre11-Rad50-ATPγS complex: Understanding the interplay between Mre11 and Rad50. Genes Dev 25: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RAM, Wittenberg C, Russell P 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Moiani D, Kertokalio A, Wyman C, Tainer JA, Russell P 2012. Mre11 ATLD17/18 mutation retains Tel1/ATM activity but blocks DNA double-strand break repair. Nucleic Acids Res 40: 11435–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Sampathi S, Dai H, Liu C, Zhou M, Hu J, Huang Q, Campbell J, Shin-Ya K, Zheng L, et al. 2013. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J 32: 1425–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R 2004. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Liu Z, Macias MJ, Bottomley MJ, Stier G, Linge JP, Nilges M, Bork P, Sattler M 1999. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure 7: 1557–1566 [DOI] [PubMed] [Google Scholar]
- Liu S, Tian LF, Liu YP, An XM, Tang Q, Yan XX, Liang DC 2014. Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair. Acta Crystallogr D Biol Crystallogr 70: 299–309 [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS 2004. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ 2009. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell 139: 100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193 [DOI] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M 2009. DNA double-strand breaks in meiosis: Checking their formation, processing and repair. DNA Repair (Amst) 8: 1127–1138 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci 96: 7376–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Alford B, Ausio J, Finn RM, McMurray CT 2012. ATP hydrolysis by RAD50 protein switches MRE11 enzyme from endonuclease to exonuclease. J Biol Chem 287: 2328–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthei KA, Keck JL 2013. The BLM dissolvasome in DNA replication and repair. Cell Mol Life Sci 70: 4067–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzan A, Pfeiffer G, Hefferin ML, Lang CE, Carney JP, Hopfner K-P 2004. MlaA, a hexameric ATPase linked to the Mre11 complex in archaeal genomes. EMBO Rep 5: 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Zinkel R, Petrini JH 2001. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet 27: 417–421 [DOI] [PubMed] [Google Scholar]
- Matsui E, Musti KV, Abe J, Yamasaki K, Matsui I, Harata K 2002. Molecular structure and novel DNA binding sites located in loops of flap endonuclease-1 from Pyrococcus horikoshii. J Biol Chem 277: 37840–37847 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Miyamoto T, Sakamoto H, Izumi H, Nakazawa Y, Ogi T, Tahara H, Oku S, Hiramoto A, Shiiki T, et al. 2011. Two unrelated patients with MRE11A mutations and Nijmegen breakage syndrome-like severe microcephaly. DNA Repair (Amst) 10: 314–321 [DOI] [PubMed] [Google Scholar]
- McKee AH, Kleckner N 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146: 797–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Mehta A, Haber JE 2014. Sources of DNA double-stand breaks and models for recombinational DNA repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J 2008. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol 181: 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29: 3358–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto R, Morino H, Yoshizawa A, Miyazaki Y, Maruyama H, Murakami N, Fukada K, Izumi Y, Matsuura S, Kaji R, et al. 2013. Exome sequencing reveals a novel MRE11 mutation in a patient with progressive myoclonic ataxia. J Neurol Sci 337: 219–223 [DOI] [PubMed] [Google Scholar]
- Möckel C, Lammens K, Schele A, Hopfner KP 2012. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res 40: 914–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C 2005. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437: 440–443 [DOI] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ 2005. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol 25: 4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD 2001a. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD 2001b. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408 [DOI] [PubMed] [Google Scholar]
- Nakada D, Matsumoto K, Sugimoto K 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev 17: 1957–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT 2010. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol 17: 1478–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Özsoy AZ, Genschel J, Modrich P, Kowalczykowski SC 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci 105: 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W 2005. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell 121: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, Beese LS 2011. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 145: 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri S, Rufa A, Pucci B, Santarnecchi E, Malandrini A, Stromillo ML, Mandala M, Rosini F, De Stefano N, Federico A 2013. Clinical course of two Italian siblings with ataxia-telangiectasia-like disorder. Cerebellum 12: 596–599 [DOI] [PubMed] [Google Scholar]
- Park YB, Chae J, Kim YC, Cho Y 2011. Crystal structure of human Mre11: Understanding tumorigenic mutations. Structure 19: 1591–1602 [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979 [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev 13: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino S, Radzimanowski J, de Sanctis D, Boeri Erba E, McSweeney S, Timmins J 2012. Structural and functional characterization of an SMC-like protein RecN: New insights into double-strand break repair. Structure 20: 2076–2089 [DOI] [PubMed] [Google Scholar]
- Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J 1996. Crystal structures of human DNA polymerase β complexed with DNA: Implications for catalytic mechanism, processivity, and fidelity. Biochemistry 35: 12742–12761 [DOI] [PubMed] [Google Scholar]
- Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, Cooper PK, Chen DJ, Tainer JA 2006. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol 13: 414–422 [DOI] [PubMed] [Google Scholar]
- Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, Vindigni A, Gileadi O 2009. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci 106: 1039–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts SA, Kullar HS, Stankovic T, Stewart GS, Last JI, Bedenham T, Armstrong SJ, Piane M, Chessa L, Taylor AM, et al. 2001. hMRE11: Genomic structure and a null mutation identified in a transcript protected from nonsense-mediated mRNA decay. Hum Mol Genet 10: 1155–1162 [DOI] [PubMed] [Google Scholar]
- Pokharel S, Campbell JL 2012. Cross talk between the nuclease and helicase activities of Dna2: Role of an essential iron-sulfur cluster domain. Nucleic Acids Res 40: 7821–7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Amon A, Klein F 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146: 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaiser A, Constantinesco F, White MF, Forterre P, Elie C 2008. The Mre11 protein interacts with both Rad50 and the HerA bipolar helicase and is recruited to DNA following γ-irradiation in the archaeon Sulfolobus acidocaldarius. BMC Mol Biol 9: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis CC, Batista S, Ferreira MG 2012. The fission yeast MRN complex tethers dysfunctional telomeres for NHEJ repair. EMBO J 31: 4576–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, Hakoshima T 2005. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J 24: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Mishima M, Nagai A, Kim SY, Ito Y, Hakoshima T, Jee JG, Kitano K 2010. Solution structure of the HRDC domain of human Bloom syndrome protein BLM. J Biochem 148: 517–525 [DOI] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. 1995. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268: 1749–1753 [DOI] [PubMed] [Google Scholar]
- Schaeper U, Subramanian T, Lim L, Boyd JM, Chinnadurai G 1998. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem 273: 8549–8552 [DOI] [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Möckel C, Schele A, Strasser K, Jackson SP, et al. 2012. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol 19: 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R 1998. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res 58: 4537–4442 [PubMed] [Google Scholar]
- Shen B, Nolan JP, Sklar LA, Park MS 1997. Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res 25: 3332–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, et al. 2014. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell 53: 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M 2008. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol 181: 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99: 577–587 [DOI] [PubMed] [Google Scholar]
- Stracker TH, Morales M, Couto SS, Hussein H, Petrini JH 2007. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature 447: 218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland BM, Bennett PV, Sidorkina O, Laval J 2000. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc Natl Acad Sci 97: 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan MK, Legris V, Tanner A, Reaper M, Vial S, Bordas R, Pollard JR, Charlton PA, Golec JMC, Bertrand A 2014. Structure of human Bloom's syndrome helicase in complex with ADP and duplex DNA. Acta Cryst 70: 1465–1475 [DOI] [PubMed] [Google Scholar]
- *.Symington LS 2014. End resection at DNA double-strand breaks: Mechanism and regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P, Smith GR 1992. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem 267: 3014–3023 [PubMed] [Google Scholar]
- Szankasi P, Smith GR 1995. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267: 1166–1169 [DOI] [PubMed] [Google Scholar]
- Tauchi H, Kobayashi J, Morishima K, Matsuura S, Nakamura A, Shiraishi T, Ito E, Masnada D, Delia D, Komatsu K 2001. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50-hMRE11-NBS1 complex DNA repair activity. J Biol Chem 276: 12–15 [DOI] [PubMed] [Google Scholar]
- Thompson LH 2012. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: The molecular choreography. Mutat Res 751: 158–246 [DOI] [PubMed] [Google Scholar]
- Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD 1997. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci 94: 7487–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD 1998. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res 58: 5027–5031 [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V 2006. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Sung P 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50-Mre11 complex. J Biol Chem 276: 35458–35464 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Yuan SS, Lee EY, Sung P 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and 95. J Biol Chem 273: 21447–21450 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Roh DH, Chen L, Van Komen S, Tomkinson A, Sung P 2003. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J Biol Chem 278: 48957–48964 [DOI] [PubMed] [Google Scholar]
- Truong LN, Li Y, Shi LZ, Hwang PY, He J, Wang H, Razavian N, Berns MW, Wu X 2013. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci 110: 7720–7725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 11: 2221–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Mitsuoka C, Terasawa M, Ogawa H, Ogawa T 2005. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol Biol Cell 16: 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Tainer JA 2012. Double strand binding-single strand incision mechanism for human flap endonuclease: Implications for the superfamily. Mech Ageing Dev 133: 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. 2011. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145: 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchisaka N, Takahashi N, Sato M, Kikuchi A, Mochizuki S, Imai K, Nonoyama S, Ohara O, Watanabe F, Mizutani S, et al. 2009. Two brothers with ataxia-telangiectasia-like disorder with lung adenocarcinoma. J Pediatr 155: 435–438 [DOI] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95: 705–716 [DOI] [PubMed] [Google Scholar]
- van Noort J, van Der Heijden T, de Jager M, Wyman C, Kanaar R, Dekker C (2003). The coiled-coil of the human Rad50 DNA repair protein contains specific segments of increased flexibility. Proc Natl Acad Sci 100: 7581–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, et al. 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93: 467–476 [DOI] [PubMed] [Google Scholar]
- Vindigni A, Hickson ID 2009. RecQ helicases: Multiple structures for multiple functions? HFSP J 3: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindigni A, Marino F, Gileadi O 2010. Probing the structural basis of RecQ helicase function. Biophys Chem 149: 67–77 [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412: 607–614 [DOI] [PubMed] [Google Scholar]
- Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, et al. 2009. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet 84: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 14: 927–939 [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ 2009. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461: 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shao Z, Shi LZ, Hwang PY, Truong LN, Berns MW, Chen DJ, Wu X 2012. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J Biol Chem 287: 21471–21480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, Zhang S, Zhu S, Sheng D, Ni J, Shen Y 2008. Physical and functional interaction between archaeal single-stranded DNA-binding protein and the 5′-3′ nuclease NurA. Biochem Biophys Res Commun 367: 523–529 [DOI] [PubMed] [Google Scholar]
- Wei T, Zhang S, Hou L, Ni J, Sheng D, Shen Y 2011. The carboxyl terminal of the archaeal nuclease NurA is involved in the interaction with single-stranded DNA-binding protein and dimer formation. Extremophiles 15: 227–234 [DOI] [PubMed] [Google Scholar]
- Westmoreland JW, Resnick MA 2013. Coincident resection at both ends of random, γ-induced double-strand breaks requires MRX (MRN), Sae2 (Ctp1), and Mre11-nuclease. PLoS Genet 9: e1003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley DB 2013. Bacterial DNA repair: Recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat Rev Microbiol 11: 9–13 [DOI] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. 2009. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM III, Carney JP, Coleman MA, Adamson AW, Christensen M, Lamerdin JE 1998. Hex1: A new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic Acids Res 26: 3762–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltzius JJ, Hohl M, Fleming JC, Petrini JH 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol 12: 403–407 [DOI] [PubMed] [Google Scholar]
- Wong AK, Ormonde PA, Pero R, Chen Y, Lian L, Salada G, Berry S, Lawrence Q, Dayananth P, Ha P, et al. 1998. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene 17: 2279–2285 [DOI] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J 2008. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci 105: 11200–11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Takai H, De Lange T 2012. Telomeric 3′ overhangs derive from resection by Exo1 and apollo and fill-in by POT1b-associated CST. Cell 150: 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YN, Bazeille N, Ding XY, Lu XM, Wang PY, Bugnard E, Grondin V, Dou SX, Xi XG 2012a. Multimeric BLM is dissociated upon ATP hydrolysis and functions as monomers in resolving DNA structures. Nucleic Acids Res 40: 9802–9814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zan H, Pone EJ, Mai T, Casali P 2012b. Immunoglobulin class-switch DNA recombination: Induction, targeting and beyond. Nat Rev Immunol 12: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ratcliff GC, Wang H, Davis-Searles PR, Gray MD, Erie DA, Redinbo MR 2002. A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3′-5′ exonuclease and 5′-protruding strand endonuclease activities. Biochemistry 41: 2901–2912 [DOI] [PubMed] [Google Scholar]
- Yang W, Hendrickson WA, Crouch RJ, Satow Y 1990. Structure of ribonuclease H phased at 2 Å resolution by MAD analysis of the selenomethionyl protein. Science 249: 1398–1405 [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Cammack R, Dillingham MS 2009. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J Biol Chem 284: 7746–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Claverie JM, Ogata H 2011. Mimivirus reveals Mre11/Rad50 fusion proteins with a sporadic distribution in eukaryotes, bacteria, viruses and plasmids. Virol J 8: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25: 5363–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem 273: 25388–25392 [DOI] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J 2003. The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Yuan J, Chen J 2009. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J Biol Chem 284: 31746–31752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahra R, Blackwood JK, Sales J, Leach DR 2007. Proofreading and secondary structure processing determine the orientation dependence of CAG-CTG trinucleotide repeat instability in Escherichia coli. Genetics 176: 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wei T, Hou G, Zhang C, Liang P, Ni J, Sheng D, Shen Y 2008. Archaeal DNA helicase HerA interacts with Mre11 homologue and unwinds blunt-ended double-stranded DNA and recombination intermediates. DNA Repair 7: 380–391 [DOI] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A 2001. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol 11: 105–109 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zickler D, Kleckner N 2014. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]