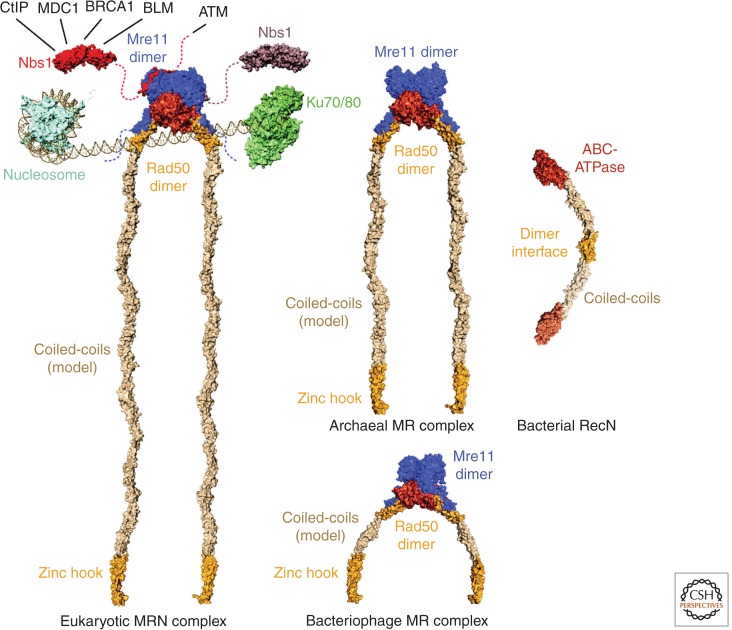
Figure 1.
The Mre11-Rad50-Nbs1 complex and phylogenetic orthologs. Structural model of MR(N) complexes together with a nucleosome, the Ku-DNA complex and RecN. Nbs1 interaction partners are indicated. The eukaryotic MRN model was built from Schizosaccharomyces pombe MN and Nbs1 (PDB code 4FBW, Schiller et al. 2012), Methanocaldococcus jannaschii MR, Pyrococcus furiosus Zn-hook and a coiled-coil model. The archaeal model is based on the M. jannaschii MR structure and the P. furiosus Zn-hook. Bacteriophage MR is modeled on the Thermotoga maritima MR complex together with the P. furiosus Zn-hook and a coiled-coil model. PDB codes are 1AOI (nucleosome, Luger et al. 1997), 1JEY (Ku-DNA complex, Walker et al. 2001), 4AD8 and 4ABX (RecN, Pellegrino et al. 2012), 4FBW (MN complex, Schiller et al. 2012), 3HUE (Nbs1, Williams et al. 2009), 3AVO (MR complex, Lim et al. 2011), and 1L8D (Zn-hook, Hopfner et al. 2002).