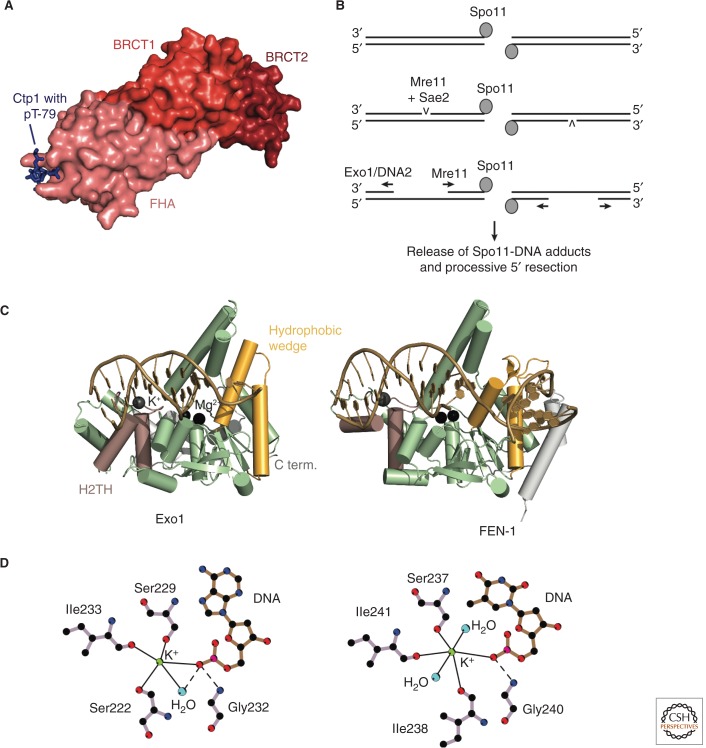
Figure 4.
Resection initiation and the Exo1 resection pathway. (A) Structure of an Nbs1-Ctp1 complex from S. pombe (PDB code is 3HUF, Williams et al. 2009). Nbs1 surface with highlighted FHA (salmon), BRCT1 (red), and BRCT2 (dark red) domains. Peptide from Ctp1 (blue) with phosphorylated T79 bound to the FHA domain. (B) Model for bidirectional resection at meiotic DSBs by Mre11 and Exo1/Sgs1-Dna2. A study of meiotic resection in S. cerevisiae suggests an endonucleotic cleavage of the 5′ strand by Mre11/Sae2 at a distance of up to 300 bp away from the Spo11-blocked DNA end. The nicked DNA can then be processed bidirectionally by Mre11 in the 3′-5′ direction and by Exo1 or Sgs1-Dna2 in the 5′-3′ direction (Garcia et al. 2011). (C) Comparison of human Exo1 and FEN-1. Features discussed in the text are highlighted and labeled for Exo1. The PDB codes are 3QEA (Exo1, Orans et al. 2011) and 3Q8K (FEN-1, Tsutakawa et al. 2011). (D) The helix-two-turn-helix (H2TH)-K+ motif in Exo1 (left) and FEN-1 (right). Straight lines indicate metal coordination, and dashed lines indicate hydrogen bonds. Only residues involved in K+-coordination and binding of the K+-coordinating DNA base are shown. The scheme was drawn using LIGPLOT (Laskowski and Swindells 2011). C term., carboxy terminal.