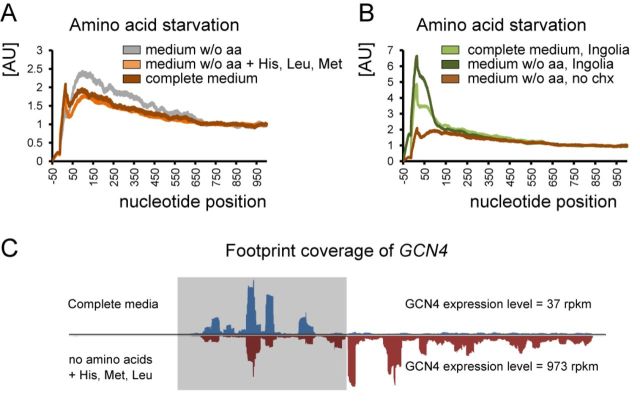
Figure 3.
In-depth investigation of amino acid starvation and changes in the ribosomal profile. (A) Repeating the experiment as was done in (1) but without cycloheximide pre-treatment still leads to a slightly different ribosomal occupancy profiles (gray and dark brown lines on the graph). However, the yeast strain BY4741, used in that study is auxotroph in histidine, leucine and methionine, which are used as selective markers. Depletion of culture medium of all amino acids cannot be considered as starvation, because the lack of three essential amino acids will lead to cell death rather than to metabolism switching toward synthesis of its own amino acids. Therefore, we supplemented the medium without amino acids with normal levels of His, Met and Leu. As a result, the difference in ribosomal profiles between starved and non-starved conditions disappeared. Thus, amino acid starvation does not cause the accumulation of ribosomes at the beginning of ORFs or uORFs. The only scenario when this accumulation was observed is the absolute lack of the essential amino acids, leading to ribosome stalling at corresponding codons. This is a very extreme case, which has little in common with regulation per se. (B) Ribo-seq data published in (1) were processed with our analytical approaches (cycloheximide was present in the culture medium). Brown line is based on our experiment (same as in Figure 3A). (C) Footprint coverage of GCN4 in response to amino acid stress derived from our experiments.