Figure 5.
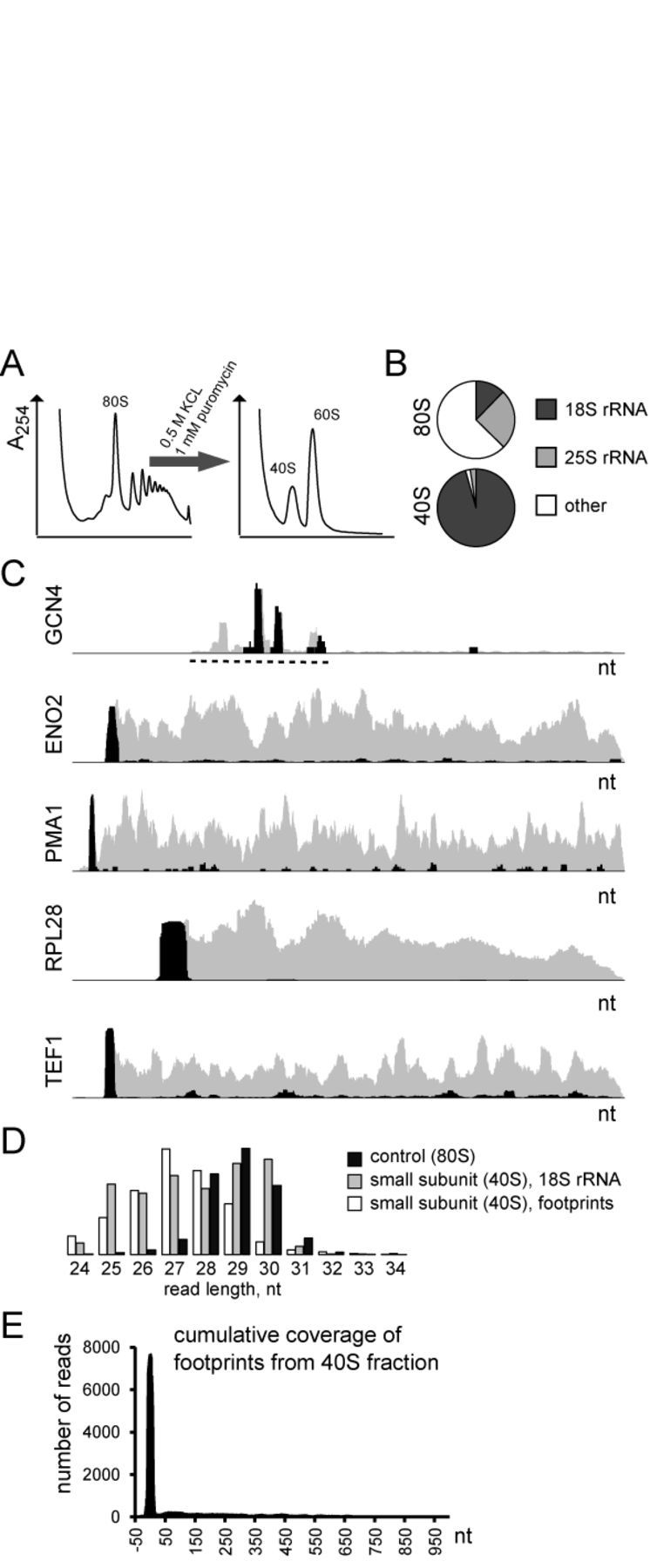
Ribo-seq of the small ribosomal subunit. (A) Dissociation of monosomes and polysomes into subunits. Fractions corresponding to the 40S small ribosomal subunit were collected for sequencing. (B) Shares of reads aligned to 18S and 25S rRNAs in Ribo-seq samples. The upper chart shows typical shares of reads in 80S fraction in control cells (YPD media, log growth phase, no stress, no cycloheximide pretreatment), and the lower shares of reads in the 40S fraction. Although not precisely quantitative, it gives an estimation of 40S fraction impurity. The ‘other’ category combines footprints and unaligned reads. (C) Representative footprint coverage profiles from normal Ribo-seq (gray) and 40S fraction (black). The dashed line marks the location of the GCN4 uORFs. (D) Footprint length distribution in 80S and 40S fractions. Reads aligned to 18S rRNA are given as a size selection control for 40S fraction. Note: each distribution class has its own scale. (E) Cumulative coverage of footprints derived from 40S fraction. No normalization was applied prior to graph plotting. All genes regardless of their length were aligned by their start codon.
