Figure 2.
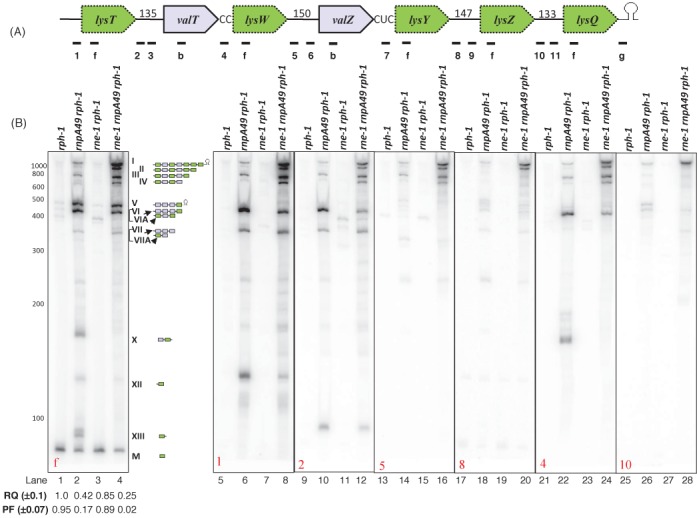
Analysis of the lysT operon processing. (A) Schematic diagram of the operon (not drawn to scale). Relative positions of the oligonucleotide probes (1: lysT-UP, f: lysmature, 2: lysT-valT1, 3: lysT-valT3, b: valUXYTZ, 4: valT-lysW, 5: lysW-valZ1, 6: valZ-UP2, 7: valZ-lysY, 8: lysY-lysZ1, 9: lysY-lysZ2, 10: lysZ-lysQ1, 11: lysZ-lysQ2, g: lysV-TER). Probe sequences are listed in Supplementary Table S1 (Supplementary Materials). Numbers indicate the length of the intergenic spacers. (B) Northern analysis of lysT operon. The specific oligonucleotide probe used in each blot is indicated in the bottom left corner of the blot. The genotypes of the strains used are indicated on the top of the each autoradiogram. The RNA molecular size standards (nucleotides) (Fermentas) are shown to the left of the first blot. The numerical designations of the different species along with their graphical structures are shown to the right of the blot. Bands VI and VII arise from the valU operon and comigrate with bands VIA and VIIA, respectively. Two independent Northern blots were run (one was probed with f, 1, 2, 5 and 8; second was probed with 4 and 10). The RQ and PF values were calculated as described in the legend to Figure 1.
