Figure 3.
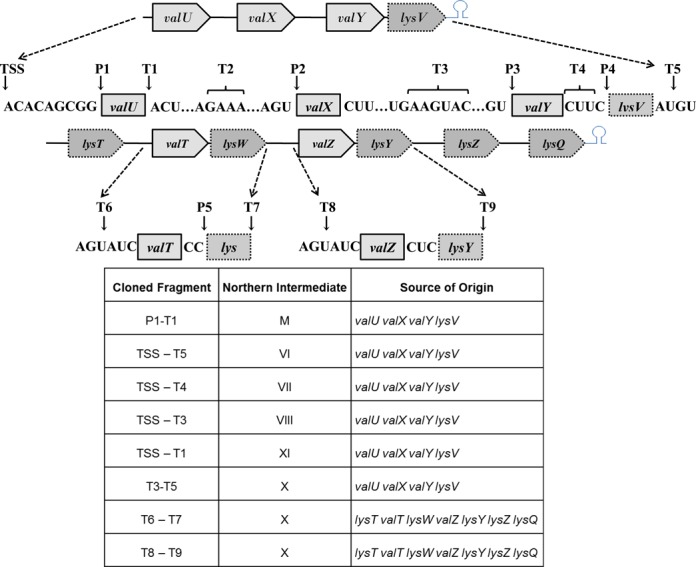
Identification of cleavage sites within the valU and lysT operons by cloning and sequencing of processing intermediates. cDNAs of self-ligated RNA molecules were amplified, cloned and sequenced to identify the 5′ and 3′ ends as described in ‘Materials and Methods’. Each arrow represents either a 5′ or 3′ end based on the DNA sequence. Multiple ends were obtained at the T2, T3 and T4 locations. The transcription, start-site (TSS), RNase P cleavage sites (P1–5), 3′ cleavage sites [either at the mature 3’ end or in the intergenic region for respective upstream tRNAs (T1–5, 7 and 9)] and 5′ cleavage sites (in the intergenic regions) for respective downstream tRNAS (T6, 8) are indicated. The 3′ and 5′ ends of some of the major processing intermediates observed in the Northern analyses (Figures 1 and 2) are shown below the diagram.
