Figure 2.
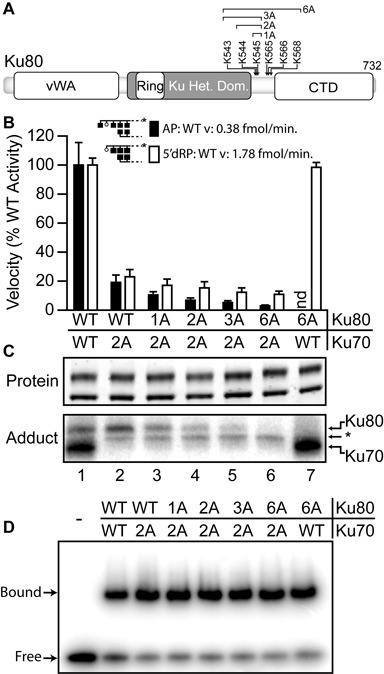
Identification of adducting lysines within human Ku80. A. Domain map of human Ku80 showing the location of lysines or groups of lysines that were mutated to alanine (A). B. Average velocities were determined from reactions at 37°C with 5 nM purified recombinant Ku heterodimer and 1 nM AP (filled bar) or 5′dRP (open bar) substrates, and expressed as a percentage of the velocity observed with WT heterodimer. Error bars are the standard deviation of triplicate determinations. C. Purified heterodimers were subjected to SDS-PAGE analysis and detected directly by SYPRO orange (top panel). Products of Schiff-base trapping assays (bottom panel) were detected by phosphorimaging after incubation of the heterodimer with radiolabeled 5′dRP substrate as in Figure 1, panel C, except reactions were supplemented with 5 mM NaBH4 and incubated for 10 min. *, unknown adducting species. D. EMSA was performed by incubating 1 nM Ku with 1 nM radiolabeled 30 bp substrate for 15 min.
