Figure 4.
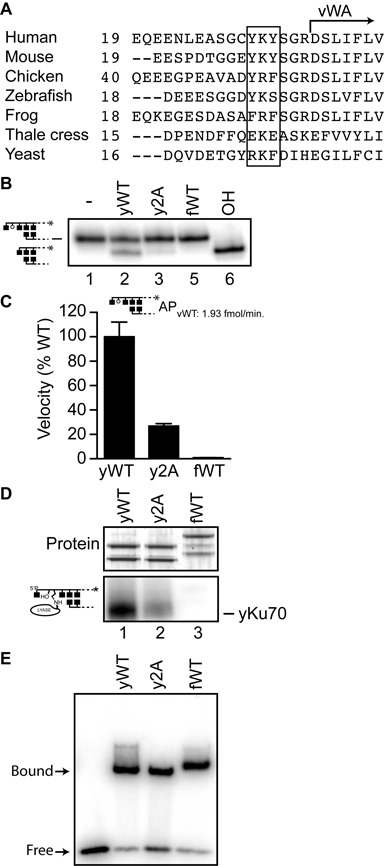
AP lyase activity of Ku from model organisms. A. Alignment of Ku70 from different species. The start of the vWA domain is noted. K31 and flanking aromatics (targeted for mutation in Figure 5) are boxed. B. Reactions were performed with 1 nM radiolabeled AP substrate with 5 nM purified recombinant Ku heterodimer at 37°C for 2.5 min, using recombinant Ku heterodimer from S. cerevisiae (yWT), a yKu heterodimer with K29A and K161A substitutions in Ku70 (y2A), or a recombinant Ku heterodimer from X. laevis (fWT). In lane 6, substrate was digested with alkali (OH) to validate abasic site generation. Reactions were analyzed by denaturing PAGE. C. Average velocities were determined from reactions at 37°C with 5 nM purified recombinant Ku heterodimer and 1 nM AP substrate, and expressed as a percentage of the velocity observed with WT yeast Ku heterodimer (yWT). Error bars are the standard deviation of triplicate determinations. D. Reactions were performed as in panel B, but supplemented with NaBH4, and analyzed after a 10 min incubation by SDS-PAGE and phosphorimaging. E. EMSA was performed by incubating 1nM Ku with 1 nM radiolabeled 30 bp substrate for 15 min.
